Nanopore unzipping of individual DNA hairpin molecules
- PMID: 15347593
- PMCID: PMC1304790
- DOI: 10.1529/biophysj.104.047274
Nanopore unzipping of individual DNA hairpin molecules
Abstract
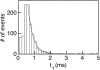
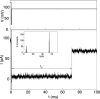
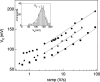
We have used the nanometer scale alpha-Hemolysin pore to study the unzipping kinetics of individual DNA hairpins under constant force or constant loading rate. Using a dynamic voltage control method, the entry rate of polynucleotides into the pore and the voltage pattern applied to induce hairpin unzipping are independently set. Thus, hundreds of unzipping events can be tested in a short period of time (few minutes), independently of the unzipping voltage amplitude. Because our method does not entail the physical coupling of the molecules under test to a force transducer, very high throughput can be achieved. We used our method to study DNA unzipping kinetics at small forces, which have not been accessed before. We find that in this regime the static unzipping times decrease exponentially with voltage with a characteristic slope that is independent of the duplex region sequence, and that the intercept depends strongly on the duplex region energy. We also present the first nanopore dynamic force measurements (time varying force). Our results are in agreement with the approximately logV dependence at high V (where V is the loading rate) observed by other methods. The extension of these measurements to lower loading rates reveals a much weaker dependence on V.
Figures




References
-
- Alberts, B., D. Bray, J. Lewis, M. Raff, K. Roberts, and J. D. Watson. 1994. Molecular Biology of the Cell. Garland Publishing, New York.
-
- Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. - PubMed
-
- Bockelmann, U., B. Essevaz-Roulet, and F. Heslot. 1997. Molecular stick-slip motion revealed by opening DNA with piconewton forces. Phys. Rev. Lett. 79:4489–4492.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

