The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling
- PMID: 15347784
- PMCID: PMC523322
- DOI: 10.1104/pp.104.046482
The potassium-dependent transcriptome of Arabidopsis reveals a prominent role of jasmonic acid in nutrient signaling
Abstract
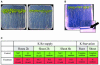
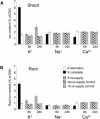
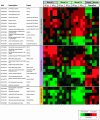
Full genome microarrays were used to assess transcriptional responses of Arabidopsis seedlings to changing external supply of the essential macronutrient potassium (K(+)). Rank product statistics and iterative group analysis were employed to identify differentially regulated genes and statistically significant coregulated sets of functionally related genes. The most prominent response was found for genes linked to the phytohormone jasmonic acid (JA). Transcript levels for the JA biosynthetic enzymes lipoxygenase, allene oxide synthase, and allene oxide cyclase were strongly increased during K(+) starvation and quickly decreased after K(+) resupply. A large number of well-known JA responsive genes showed the same expression profile, including genes involved in storage of amino acids (VSP), glucosinolate production (CYP79), polyamine biosynthesis (ADC2), and defense (PDF1.2). Our findings highlight a novel role of JA in nutrient signaling and stress management through a variety of physiological processes such as nutrient storage, recycling, and reallocation. Other highly significant K(+)-responsive genes discovered in our study encoded cell wall proteins (e.g. extensins and arabinogalactans) and ion transporters (e.g. the high-affinity K(+) transporter HAK5 and the nitrate transporter NRT2.1) as well as proteins with a putative role in Ca(2+) signaling (e.g. calmodulins). On the basis of our results, we propose candidate genes involved in K(+) perception and signaling as well as a network of molecular processes underlying plant adaptation to K(+) deficiency.
Figures


References
-
- Amtmann A, Armengaud P, Volkov V (2004) Potassium nutrition and salt stress. In MR Blatt, ed, Membrane Transport in Plants. Blackwell Publishing, Oxford (in press)
-
- Berger S (2002) Jasmonate-related mutants of Arabidopsis as tools for studying stress signaling. Planta 214: 497–504 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

