Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation
- PMID: 15347814
- PMCID: PMC518776
- DOI: 10.1073/pnas.0403851101
Distinct roles of transcription factors TFIIIB and TFIIIC in RNA polymerase III transcription reinitiation
Abstract
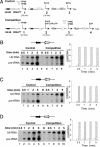
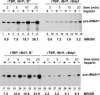
Eukaryotic RNA polymerase (Pol) III is recruited to target promoters by a stable preinitiation complex containing transcription factors TFIIIC and TFIIIB. After the first transcription cycle, reinitiation proceeds through facilitated recycling, a process by which the terminating Pol III rapidly reloads onto the same transcription unit. Here, we show that Pol III is repeatedly recaptured in vitro by the first transcribed gene, even in the presence of a juxtaposed competitor promoter complex, thus suggesting that facilitated recycling is not merely due to a stochastic reassociation process favored by the small size of class III genes. The transcription factor requirements for facilitated reinitiation were investigated by taking advantage of Pol III templates that support both TFIIIC-dependent and TFIIIC-independent transcription. A TFIIIC-less transcription system, in which TFIIIB was reconstituted from recombinant TATA box-binding protein and Brf1 proteins and a crude fraction containing the Bdp1 component, was sufficient to direct efficient Pol III recycling on short ( approximately 100 bp) class III genes. Unexpectedly, however, on longer (>300 bp) transcription units, reinitiation in the presence of TFIIIB alone was compromised, and TFIIIC was further required to reestablish a high reinitiation rate. Transcription reinitiation was also severely impaired when recombinant Bdp1 protein replaced the corresponding crude fraction in reconstituted TFIIIB. The data reveal an unexpected complexity in the Pol III reinitiation mechanism and suggest the existence of a handing-back network between Pol III, TFIIIC, and TFIIIB on actively transcribed class III genes.
Figures



References
-
- Dieci, G. & Sentenac, A. (2003) Trends. Biochem. Sci. 28, 202–209. - PubMed
-
- Geiduschek, E. P. & Kassavetis, G. A. (2001) J. Mol. Biol. 310, 1–26. - PubMed
-
- Dieci, G. & Sentenac, A. (1996) Cell 84, 245–252. - PubMed
-
- Guffanti, E., Corradini, R., Ottonello, S. & Dieci, G. (2004) J. Biol. Chem. 279, 20708–20716. - PubMed
-
- Dieci, G., Percudani, R., Giuliodori, S., Bottarelli, L. & Ottonello, S. (2000) J. Mol. Biol. 299, 601–613. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

