Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing
- PMID: 15347815
- PMCID: PMC518758
- DOI: 10.1073/pnas.0403844101
Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing
Abstract
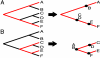
Phylogenetic reconstruction using molecular data is often subject to homoplasy, leading to inaccurate conclusions about phylogenetic relationships among operational taxonomic units. Compared with other molecular markers, single-nucleotide polymorphisms (SNPs) exhibit extremely low mutation rates, making them rare in recently emerged pathogens, but they are less prone to homoplasy and thus extremely valuable for phylogenetic analyses. Despite their phylogenetic potential, ascertainment bias occurs when SNP characters are discovered through biased taxonomic sampling; by using whole-genome comparisons of five diverse strains of Bacillus anthracis to facilitate SNP discovery, we show that only polymorphisms lying along the evolutionary pathway between reference strains will be observed. We illustrate this in theoretical and simulated data sets in which complex phylogenetic topologies are reduced to linear evolutionary models. Using a set of 990 SNP markers, we also show how divergent branches in our topologies collapse to single points but provide accurate information on internodal distances and points of origin for ancestral clades. These data allowed us to determine the ancestral root of B. anthracis, showing that it lies closer to a newly described "C" branch than to either of two previously described "A" or "B" branches. In addition, subclade rooting of the C branch revealed unequal evolutionary rates that seem to be correlated with ecological parameters and strain attributes. Our use of nonhomoplastic whole-genome SNP characters allows branch points and clade membership to be estimated with great precision, providing greater insight into epidemiological, ecological, and forensic questions.
Figures



References
-
- Read, T. D., Salzberg, S. L., Pop, M., Shumway, M., Umayam, L., Jiang, L., Holtzapple, E., Busch, J. D., Smith, K. L., Schupp, J. M., et al. (2002) Science 296, 2028–2033. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

