GASP: Gapped Ancestral Sequence Prediction for proteins
- PMID: 15350199
- PMCID: PMC517926
- DOI: 10.1186/1471-2105-5-123
GASP: Gapped Ancestral Sequence Prediction for proteins
Abstract
Background: The prediction of ancestral protein sequences from multiple sequence alignments is useful for many bioinformatics analyses. Predicting ancestral sequences is not a simple procedure and relies on accurate alignments and phylogenies. Several algorithms exist based on Maximum Parsimony or Maximum Likelihood methods but many current implementations are unable to process residues with gaps, which may represent insertion/deletion (indel) events or sequence fragments.
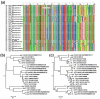
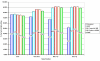
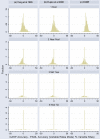
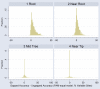
Results: Here we present a new algorithm, GASP (Gapped Ancestral Sequence Prediction), for predicting ancestral sequences from phylogenetic trees and the corresponding multiple sequence alignments. Alignments may be of any size and contain gaps. GASP first assigns the positions of gaps in the phylogeny before using a likelihood-based approach centred on amino acid substitution matrices to assign ancestral amino acids. Important outgroup information is used by first working down from the tips of the tree to the root, using descendant data only to assign probabilities, and then working back up from the root to the tips using descendant and outgroup data to make predictions. GASP was tested on a number of simulated datasets based on real phylogenies. Prediction accuracy for ungapped data was similar to three alternative algorithms tested, with GASP performing better in some cases and worse in others. Adding simple insertions and deletions to the simulated data did not have a detrimental effect on GASP accuracy.
Conclusions: GASP (Gapped Ancestral Sequence Prediction) will predict ancestral sequences from multiple protein alignments of any size. Although not as accurate in all cases as some of the more sophisticated maximum likelihood approaches, it can process a wide range of input phylogenies and will predict ancestral sequences for gapped and ungapped residues alike.
Figures
References
-
- Zhang J, Nei M. Accuracies of ancestral amino acid sequences inferred by the parsimony, likelihood, and distance methods. J Mol Evol. 1997;44:S139–146. - PubMed
-
- Fitch WM. Toward defining course of evolution – minimum change for a specific tree topology. Systematic Zoology. 1971;20:406–416.
-
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

