Protein-polymer nano-machines. Towards synthetic control of biological processes
- PMID: 15350203
- PMCID: PMC519025
- DOI: 10.1186/1477-3155-2-8
Protein-polymer nano-machines. Towards synthetic control of biological processes
Abstract
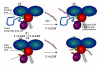
The exploitation of nature's machinery at length scales below the dimensions of a cell is an exciting challenge for biologists, chemists and physicists, while advances in our understanding of these biological motifs are now providing an opportunity to develop real single molecule devices for technological applications. Single molecule studies are already well advanced and biological molecular motors are being used to guide the design of nano-scale machines. However, controlling the specific functions of these devices in biological systems under changing conditions is difficult. In this review we describe the principles underlying the development of a molecular motor with numerous potential applications in nanotechnology and the use of specific synthetic polymers as prototypic molecular switches for control of the motor function. The molecular motor is a derivative of a TypeI Restriction-Modification (R-M) enzyme and the synthetic polymer is drawn from the class of materials that exhibit a temperature-dependent phase transition.The potential exploitation of single molecules as functional devices has been heralded as the dawn of new era in biotechnology and medicine. It is not surprising, therefore, that the efforts of numerous multidisciplinary teams 12. have been focused in attempts to develop these systems. as machines capable of functioning at the low sub-micron and nanometre length-scales 3. However, one of the obstacles for the practical application of single molecule devices is the lack of functional control methods in biological media, under changing conditions. In this review we describe the conceptual basis for a molecular motor (a derivative of a TypeI Restriction-Modification enzyme) with numerous potential applications in nanotechnology and the use of specific synthetic polymers as prototypic molecular switches for controlling the motor function 4.
Figures




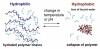
References
-
- Firman K, Dutta C, Weiserova M, Janscak P. The role of subunit assembly in the functional control of type I restriction-modification enzymes. Molecular Biology Today. 2000;1:1–8.
Grants and funding
LinkOut - more resources
Full Text Sources

