In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine
- PMID: 15351731
- PMCID: PMC7092815
- DOI: 10.1016/j.bbrc.2004.08.085
In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine
Abstract
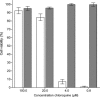
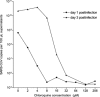
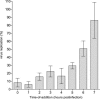
We report on chloroquine, a 4-amino-quinoline, as an effective inhibitor of the replication of the severe acute respiratory syndrome coronavirus (SARS-CoV) in vitro. Chloroquine is a clinically approved drug effective against malaria. We tested chloroquine phosphate for its antiviral potential against SARS-CoV-induced cytopathicity in Vero E6 cell culture. Results indicate that the IC50 of chloroquine for antiviral activity (8.8 +/- 1.2 microM) was significantly lower than its cytostatic activity; CC50 (261.3 +/- 14.5 microM), yielding a selectivity index of 30. The IC50 of chloroquine for inhibition of SARS-CoV in vitro approximates the plasma concentrations of chloroquine reached during treatment of acute malaria. Addition of chloroquine to infected cultures could be delayed for up to 5h postinfection, without an important drop in antiviral activity. Chloroquine, an old antimalarial drug, may be considered for immediate use in the prevention and treatment of SARS-CoV infections.
Copyright 2004 Elsevier Inc.
Figures
References
-
- World Health Organization, 2003. Communicable Disease Surveillance and Response. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. Available from: <http://www.who.int/csr/sars/country/table2003_09_23/en/>. (revised 26 September 2003)
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J., SARS Working Group A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Kuiken T., Fouchier R.A., Schutten M., Rimmelzwaan G.F., van Amerongen G., van Riel D., Laman J.D., de Jong T., van Doornum G., Lim W., Ling A.E., Chan P.K., Tam J.S., Zambon M.C., Gopal R., Drosten C., van der Werf S., Escriou N., Manuguerra J.C., Stohr K., Peiris J.S., Osterhaus A.D. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362:263–270. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

