A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus
- PMID: 15353561
- PMCID: PMC519124
- DOI: 10.1093/nar/gnh122
A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus
Abstract
Altered methylation patterns have been found to play a role in developmental disorders, cancer and aging. Increasingly, changes in DNA methylation are used as molecular markers of disease. Therefore, there is a need for reliable and easy to use techniques to detect and measure DNA methylation in research and routine diagnostics. We have established a novel quantitative analysis of methylated alleles (QAMA) which is essentially a major improvement over a previous method based on real-time PCR (MethyLight). This method is based on real-time PCR on bisulfite-treated DNA. A significant advantage over conventional MethyLight is gained by the use of TaqMan probes based on minor groove binder (MGB) technology. Their improved sequence specificity facilitates relative quantification of methylated and unmethylated alleles that are simultaneously amplified in single tube. This improvement allows precise measurement of the ratio of methylated versus unmethylated alleles and cuts down potential sources of inter-assay variation. Therefore, fewer control assays are required. We have used this novel technical approach to identify hypermethylation of the CpG island located in the promoter region of the retinoblastoma (RB1) gene and found that QAMA facilitates reliable and fast measurement of the relative quantity of methylated alleles and improves handling of diagnostic methylation analysis. Moreover, the simplified reaction setup and robustness inherent to the single tube assay facilitates high-throughput methylation analysis. Because the high sequence specificity inherent to the MGB technology is widely used to discriminate single nucleotide polymorphisms, QAMA potentially can be used to discriminate the methylation status of single CpG dinucleotides.
Figures
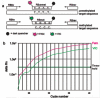

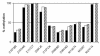
References
-
- Zeschnigk M., Lich,C., Buiting,K., Doerfler,W. and Horsthemke,B. (1997) A single-tube PCR test for the diagnosis of Angelman and Prader–Willi syndrome based on allelic methylation differences at the SNRPN locus. Eur. J. Hum. Genet., 5, 94–98. - PubMed
-
- Oberle I., Rousseau,F., Heitz,D., Kretz,C., Devys,D., Hanauer,A., Boue,J., Bertheas,M.F. and Mandel,J.L. (1991) Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science, 252, 1097–1102. - PubMed
-
- Dittrich B., Robinson,W.P., Knoblauch,H., Buiting,K., Schmidt,K., Gillessen-Kaesbach,G. and Horsthemke,B. (1992) Molecular diagnosis of the Prader–Willi and Angelman syndromes by detection of parent-of-origin specific DNA methylation in 15q11–13. Hum. Genet., 90, 313–315. - PubMed
-
- Issa J.P. (2003) Age-related epigenetic changes and the immune system. Clin. Immunol., 109, 103–108. - PubMed
-
- Jaenisch R. and Bird,A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature Genet., 33 (Suppl), 245–254. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

