Effects of length and location on the cellular response to double-stranded RNA
- PMID: 15353564
- PMCID: PMC515255
- DOI: 10.1128/MMBR.68.3.432-452.2004
Effects of length and location on the cellular response to double-stranded RNA
Abstract
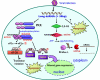
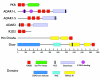
Since double-stranded RNA (dsRNA) has not until recently generally been thought to be deliberately expressed in cells, it has commonly been assumed that the major source of cellular dsRNA is viral infections. In this view, the cellular responses to dsRNA would be natural and perhaps ancient antiviral responses. While the cell may certainly react to some dsRNAs as an antiviral response, this does not represent the only response or even, perhaps, the major one. A number of recent observations have pointed to the possibility that dsRNA molecules are not seen only as evidence of viral infection or recognized for degradation because they cannot be translated. In some instances they may also play important roles in normal cell growth and function. The purpose of this review is to outline our current understanding of the fate of dsRNA in cells, with a focus on the apparent fact that their fates and functions appear to depend critically not only on where in the cell dsRNA molecules are found, but also on how long they are and perhaps on how abundant they are.
Figures



Similar articles
-
Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes.Microbiol Mol Biol Rev. 1998 Dec;62(4):1415-34. doi: 10.1128/MMBR.62.4.1415-1434.1998. Microbiol Mol Biol Rev. 1998. PMID: 9841677 Free PMC article. Review.
-
Functions of double-stranded RNA-binding domains in nucleocytoplasmic transport.RNA Biol. 2014;11(10):1226-32. doi: 10.4161/15476286.2014.972856. RNA Biol. 2014. PMID: 25584639 Free PMC article. Review.
-
Double-Stranded RNA Is Detected by Immunofluorescence Analysis in RNA and DNA Virus Infections, Including Those by Negative-Stranded RNA Viruses.J Virol. 2015 Sep;89(18):9383-92. doi: 10.1128/JVI.01299-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136565 Free PMC article.
-
Short hairpin type of dsRNAs that are controlled by tRNA(Val) promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells.Nucleic Acids Res. 2003 Jan 15;31(2):700-7. doi: 10.1093/nar/gkg158. Nucleic Acids Res. 2003. PMID: 12527779 Free PMC article.
-
Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants.New Phytol. 2016 Aug;211(3):1008-19. doi: 10.1111/nph.13944. Epub 2016 Mar 31. New Phytol. 2016. PMID: 27030513
Cited by
-
Dicer represses the interferon response and the double-stranded RNA-activated protein kinase pathway in mouse embryonic stem cells.J Biol Chem. 2021 Jan-Jun;296:100264. doi: 10.1016/j.jbc.2021.100264. Epub 2021 Jan 8. J Biol Chem. 2021. PMID: 33837743 Free PMC article.
-
Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses.J Virol. 2006 May;80(10):5059-64. doi: 10.1128/JVI.80.10.5059-5064.2006. J Virol. 2006. PMID: 16641297 Free PMC article.
-
Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells.Nucleic Acids Res. 2006;34(17):4801-15. doi: 10.1093/nar/gkl646. Epub 2006 Sep 13. Nucleic Acids Res. 2006. PMID: 16971455 Free PMC article.
-
A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L.RNA. 2008 Jun;14(6):1026-36. doi: 10.1261/rna.958908. Epub 2008 Apr 21. RNA. 2008. PMID: 18426919 Free PMC article.
-
Transfection of microRNA Mimics Should Be Used with Caution.Front Genet. 2015 Dec 2;6:340. doi: 10.3389/fgene.2015.00340. eCollection 2015. Front Genet. 2015. PMID: 26697058 Free PMC article.
References
-
- Adelman, Z. N., I. Sanchez-Vargas, E. A. Travanty, J. O. Carlson, B. J. Beaty, C. D. Blair, and K. E. Olson. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 76:12925-12933. - PMC - PubMed
-
- Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-kappa B by Toll-like receptor 3. Nature 413:732-738. - PubMed
-
- Allshire, R. 2002. Molecular biology. RNAi and heterochromatin-a hushed-up affair. Science 297:1818-1819. - PubMed
-
- Ambros, V. 2003. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113:673-676. - PubMed
-
- Ambros, V. 2001. MicroRNAs: tiny regulators with great potential. Cell 107:823-826. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

