Determination of the core of a minimal bacterial gene set
- PMID: 15353568
- PMCID: PMC515251
- DOI: 10.1128/MMBR.68.3.518-537.2004
Determination of the core of a minimal bacterial gene set
Abstract
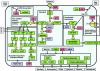
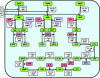
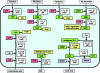
The availability of a large number of complete genome sequences raises the question of how many genes are essential for cellular life. Trying to reconstruct the core of the protein-coding gene set for a hypothetical minimal bacterial cell, we have performed a computational comparative analysis of eight bacterial genomes. Six of the analyzed genomes are very small due to a dramatic genome size reduction process, while the other two, corresponding to free-living relatives, are larger. The available data from several systematic experimental approaches to define all the essential genes in some completely sequenced bacterial genomes were also considered, and a reconstruction of a minimal metabolic machinery necessary to sustain life was carried out. The proposed minimal genome contains 206 protein-coding genes with all the genetic information necessary for self-maintenance and reproduction in the presence of a full complement of essential nutrients and in the absence of environmental stress. The main features of such a minimal gene set, as well as the metabolic functions that must be present in the hypothetical minimal cell, are discussed.
Figures
Similar articles
-
A minimal gene set for cellular life derived by comparison of complete bacterial genomes.Proc Natl Acad Sci U S A. 1996 Sep 17;93(19):10268-73. doi: 10.1073/pnas.93.19.10268. Proc Natl Acad Sci U S A. 1996. PMID: 8816789 Free PMC article.
-
Evolution of minimal-gene-sets in host-dependent bacteria.Trends Microbiol. 2004 Jan;12(1):37-43. doi: 10.1016/j.tim.2003.11.006. Trends Microbiol. 2004. PMID: 14700550 Review.
-
The minimal genome concept.Curr Opin Genet Dev. 1999 Dec;9(6):709-14. doi: 10.1016/s0959-437x(99)00023-4. Curr Opin Genet Dev. 1999. PMID: 10607608 Review.
-
Exploration and grading of possible genes from 183 bacterial strains by a common protocol to identification of new genes: Gene Trek in Prokaryote Space (GTPS).DNA Res. 2006 Dec 31;13(6):245-54. doi: 10.1093/dnares/dsl014. Epub 2006 Dec 13. DNA Res. 2006. PMID: 17166861
-
Complete genome sequences of cellular life forms: glimpses of theoretical evolutionary genomics.Curr Opin Genet Dev. 1996 Dec;6(6):757-62. doi: 10.1016/s0959-437x(96)80032-3. Curr Opin Genet Dev. 1996. PMID: 8994848 Review.
Cited by
-
Phylogeny and population structure of brown rot- and Moko disease-causing strains of Ralstonia solanacearum phylotype II.Appl Environ Microbiol. 2012 Apr;78(7):2367-75. doi: 10.1128/AEM.06123-11. Epub 2012 Jan 27. Appl Environ Microbiol. 2012. PMID: 22286995 Free PMC article.
-
Physical Routes to Primitive Cells: An Experimental Model Based on the Spontaneous Entrapment of Enzymes inside Micrometer-Sized Liposomes.Life (Basel). 2015 Mar 18;5(1):969-96. doi: 10.3390/life5010969. Life (Basel). 2015. PMID: 25793278 Free PMC article.
-
Protocells: at the interface of life and non-life.Life (Basel). 2015 Feb 9;5(1):447-58. doi: 10.3390/life5010447. Life (Basel). 2015. PMID: 25809963 Free PMC article.
-
Sinorhizobium meliloti YbeY is a zinc-dependent single-strand specific endoribonuclease that plays an important role in 16S ribosomal RNA processing.Nucleic Acids Res. 2020 Jan 10;48(1):332-348. doi: 10.1093/nar/gkz1095. Nucleic Acids Res. 2020. PMID: 31777930 Free PMC article.
-
Differential annotation of tRNA genes with anticodon CAT in bacterial genomes.Nucleic Acids Res. 2006;34(20):6015-22. doi: 10.1093/nar/gkl739. Epub 2006 Oct 27. Nucleic Acids Res. 2006. PMID: 17071718 Free PMC article.
References
-
- Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. - PubMed
-
- Arigoni, F., F. Talabot, M. Peitsch, M. D. Edgerton, E. Meldrum, E. Allet, R. Fish, T. Jamotte, M. L. Curchod, and H. Loferer. 1998. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 16:851-856. - PubMed
-
- Begley, T. P., C. Kinsland, R. A. Mehl, A. Osterman, and P. Dorrestein. 2001. The biosynthesis of nicotinamide adenine dinucleotides in bacteria. Vitam. Horm. 61:103-109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

