Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex
- PMID: 15353591
- PMCID: PMC521936
- DOI: 10.1073/pnas.0404386101
Reverse transcriptase activity innate to DNA polymerase I and DNA topoisomerase I proteins of Streptomyces telomere complex
Abstract
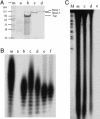
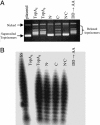
Replication of Streptomyces linear chromosomes and plasmids proceeds bidirectionally from a central origin, leaving recessed 5' termini that are extended by a telomere binding complex. This complex contains both a telomere-protecting terminal protein (Tpg) and a telomere-associated protein that interacts with Tpg and the DNA ends of linear Streptomyces replicons. By using histidine-tagged telomere-associated protein (Tap) as a scaffold, we identified DNA polymerase (PolA) and topoisomerase I (TopA) proteins as other components of the Streptomyces telomere complex. Biochemical characterization of these proteins indicated that both PolA and TopA exhibit highly efficient reverse transcriptase (RT) activity in addition to their predicted functions. Although RT activity innate to other DNA-dependent DNA polymerases has been observed previously, its occurrence in a topoisomerase is unprecedented. Deletion mapping and sequence analysis showed that the RT activity of Streptomcyces TopA resides in a peptide region containing motifs that are absent from most bacterial topoisomerases but are highly conserved in a novel subfamily of eubacterial topoisomerases found largely in Actinobacteria. Within one of these motifs, and essential to the RT function of Streptomyces TopA, is an Asp-Asp doublet sequence required also for the RT activities of human immunodeficiency virus and eukaryotic cell telomerases.
Figures

Comment in
-
Reverse transcriptase at bacterial telomeres.Proc Natl Acad Sci U S A. 2004 Oct 5;101(40):14307-8. doi: 10.1073/pnas.0406018101. Epub 2004 Sep 28. Proc Natl Acad Sci U S A. 2004. PMID: 15454610 Free PMC article. No abstract available.
References
-
- Kornberg, A. & Baker, T. A. (1992) DNA Replication (Freeman, New York), 2nd Ed.
-
- Greider, C. W. & Blackburn, E. H. (1987) Cell 51, 887-898. - PubMed
-
- Lingner, J. & Cech, T. R. (1998) Curr. Opin. Genet. Dev. 8, 226-232. - PubMed
-
- McEachern, M. J., Krauskopf, A. & Blackburn, E. H. (2000) Annu. Rev. Genet. 34, 331-358. - PubMed
-
- Blackburn, E. H. (2001) Cell 106, 661-673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

