The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae
- PMID: 15353602
- PMCID: PMC518775
- DOI: 10.1073/pnas.0405868101
The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae
Abstract
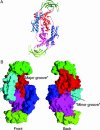
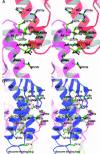
Trigger factor is a molecular chaperone that is present in all species of eubacteria. It binds to the ribosomal 50S subunit near the translation exit tunnel and is thought to be the first protein to interact with nascent polypeptides emerging from the ribosome. The chaperone has a peptidyl-prolyl cis-trans isomerase (PPIase) activity that catalyzes the rate-limiting proline isomerization in the protein-folding process. We have determined the crystal structure of nearly full-length trigger factor from Vibrio cholerae by x-ray crystallography at 2.5-A resolution. The structure is composed of two trigger-factor molecules related by a noncrystallographic two-fold symmetry axis. The monomer has an elongated shape and is folded into three domains: an N-terminal domain I that binds to the ribosome, a central domain II that contains PPIase activity, and a C-terminal domain III. The active site of the PPIase domain is occupied by a loop from domain III, suggesting that the PPIase activity of the protein could be regulated. The dimer interface is formed between domains I and III and contains residues of mixed properties. Further implications about dimerization, ribosome binding, and other functions of trigger factor are discussed.
Figures

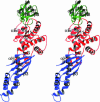

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

