Bystander gene activation by a locus control region
- PMID: 15359275
- PMCID: PMC522784
- DOI: 10.1038/sj.emboj.7600365
Bystander gene activation by a locus control region
Abstract
Random assortment of genes within mammalian genomes establishes the potential for interference between neighboring genes with distinct transcriptional specificities. Long-range transcriptional controls further increase this potential. Exploring this problem is of fundamental importance to understanding gene regulation. In the human genome, the Igbeta (CD79b) gene is situated between the pituitary-specific human growth hormone (hGH) gene and its locus control region (hGH LCR). Igbeta protein is considered B-cell specific; its only known role is in B-cell receptor signaling. Unexpectedly, we found that hIgbeta is transcribed at high levels in the pituitary. This Igbeta transcription is dependent on pituitary-specific epigenetic modifications generated by the hGH LCR. In contrast, expression of Igbeta at its native site in B cells is independent of hGH LCR activity. These studies demonstrated that a gene with tissue-restricted transcriptional determinants (B cell) can be robustly activated in an unrelated tissue (pituitary) due to fortuitous positioning within an active chromatin domain. This 'bystander' gene activation pathway impacts on current concepts of tissue specificity and models of active chromatin domains.
Figures
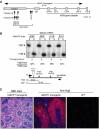
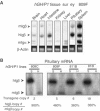



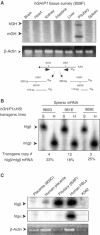

Similar articles
-
Locus control region transcription plays an active role in long-range gene activation.Mol Cell. 2006 Aug 4;23(3):365-75. doi: 10.1016/j.molcel.2006.05.041. Mol Cell. 2006. PMID: 16885026
-
Tissue-specific chromatin modifications at a multigene locus generate asymmetric transcriptional interactions.Mol Cell Biol. 2006 Aug;26(15):5569-79. doi: 10.1128/MCB.00405-06. Mol Cell Biol. 2006. PMID: 16847312 Free PMC article.
-
Expression of Pit-1 in nonsomatotrope cell lines induces human growth hormone locus control region histone modification and hGH-N transcription.J Mol Biol. 2009 Jul 3;390(1):26-44. doi: 10.1016/j.jmb.2009.04.081. Epub 2009 May 7. J Mol Biol. 2009. PMID: 19427323
-
Activation of the human GH gene cluster: roles for targeted chromatin modification.Trends Endocrinol Metab. 2004 Jan-Feb;15(1):40-5. doi: 10.1016/j.tem.2003.11.004. Trends Endocrinol Metab. 2004. PMID: 14693425 Review.
-
A family of POU-domain and Pit-1 tissue-specific transcription factors in pituitary and neuroendocrine development.Annu Rev Physiol. 1990;52:773-91. doi: 10.1146/annurev.ph.52.030190.004013. Annu Rev Physiol. 1990. PMID: 2184776 Review.
Cited by
-
Distinct chromatin configurations regulate the initiation and the maintenance of hGH gene expression.Mol Cell Biol. 2013 May;33(9):1723-34. doi: 10.1128/MCB.01166-12. Epub 2013 Feb 19. Mol Cell Biol. 2013. PMID: 23428872 Free PMC article.
-
Transcriptional interference--a crash course.Trends Genet. 2005 Jun;21(6):339-45. doi: 10.1016/j.tig.2005.04.009. Trends Genet. 2005. PMID: 15922833 Free PMC article. Review.
-
Neighbourhood continuity is not required for correct testis gene expression in Drosophila.PLoS Biol. 2010 Nov 30;8(11):e1000552. doi: 10.1371/journal.pbio.1000552. PLoS Biol. 2010. PMID: 21151342 Free PMC article.
-
Identification of a secondary promoter within the human B cell receptor component gene hCD79b.J Biol Chem. 2013 Jun 21;288(25):18353-65. doi: 10.1074/jbc.M113.461988. Epub 2013 May 6. J Biol Chem. 2013. PMID: 23649625 Free PMC article.
-
Protein-coding cis-natural antisense transcripts have high and broad expression in Arabidopsis.Plant Physiol. 2013 Apr;161(4):2171-80. doi: 10.1104/pp.112.212100. Epub 2013 Mar 1. Plant Physiol. 2013. PMID: 23457227 Free PMC article.
References
-
- Asa SL, Puy LA, Lew AM, Sundmark VC, Elsholtz HP (1993) Cell type-specific expression of the pituitary transcription activator pit-1 in the human pituitary and pituitary adenomas. J Clin Endocr Metab 77: 1275–1280 - PubMed
-
- Bell AC, Felsenfeld G (1999) Stopped at the border: boundaries insulators. Curr Opin Genet Dev 9: 191–198 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

