Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination
- PMID: 15359278
- PMCID: PMC522788
- DOI: 10.1038/sj.emboj.7600383
Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination
Abstract
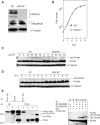
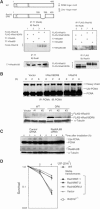
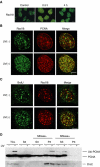
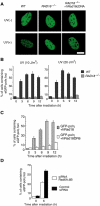
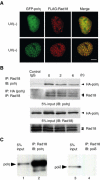
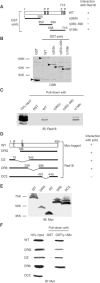
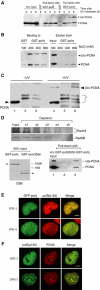
The DNA replication machinery stalls at damaged sites on templates, but normally restarts by switching to a specialized DNA polymerase(s) that carries out translesion DNA synthesis (TLS). In human cells, DNA polymerase eta (poleta) accumulates at stalling sites as nuclear foci, and is involved in ultraviolet (UV)-induced TLS. Here we show that poleta does not form nuclear foci in RAD18(-/-) cells after UV irradiation. Both Rad18 and Rad6 are required for poleta focus formation. In wild-type cells, UV irradiation induces relocalization of Rad18 in the nucleus, thereby stimulating colocalization with proliferating cell nuclear antigen (PCNA), and Rad18/Rad6-dependent PCNA monoubiquitination. Purified Rad18 and Rad6B monoubiquitinate PCNA in vitro. Rad18 associates with poleta constitutively through domains on their C-terminal regions, and this complex accumulates at the foci after UV irradiation. Furthermore, poleta interacts preferentially with monoubiquitinated PCNA, but poldelta does not. These results suggest that Rad18 is crucial for recruitment of poleta to the damaged site through protein-protein interaction and PCNA monoubiquitination.
Figures
References
-
- Bailly V, Lamb J, Sung P, Prakash S, Prakash L (1994) Specific complex formation between yeast RAD6 and RAD18 proteins: a potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev 8: 811–820 - PubMed
-
- Bailly V, Lauder S, Prakash S, Prakash L (1997a) Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem 272: 23360–23365 - PubMed
-
- Broomfield S, Hryciw T, Xiao W (2001) DNA postreplication repair and mutagenesis in Saccharomyces cerevisiae. Mutat Res 486: 167–184 - PubMed
-
- Broughton BC, Cordonnier A, Kleijer WJ, Jaspers NGJ, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF, Boudsocq F, Masutani C, Hanaoka F, Fuchs RP, Sarasin A, Lehmann AR (2002) Molecular analysis of mutation in DNA polymerase η in xeroderma pigmentosum-variant patients. Proc Natl Acad Sci USA 99: 815–820 - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

