Requirement of IS911 replication before integration defines a new bacterial transposition pathway
- PMID: 15359283
- PMCID: PMC522794
- DOI: 10.1038/sj.emboj.7600395
Requirement of IS911 replication before integration defines a new bacterial transposition pathway
Abstract
Movement of transposable elements is often accompanied by replication to ensure their proliferation. Replication is associated with both major classes of transposition mechanisms: cut-and-paste and cointegrate formation (paste-and-copy). Cut-and-paste transposition is often activated by replication of the transposon, while in cointegrate formation replication completes integration. We describe a novel transposition mechanism used by insertion sequence IS911, which we call copy-and-paste. IS911 transposes using a circular intermediate (circle), which then integrates into a target. We demonstrate that this is derived from a branched intermediate (figure-eight) in which both ends are joined by a single-strand bridge after a first-strand transfer. In vivo labelling experiments show that the process of circle formation is replicative. The results indicate that the replication pathway not only produces circles from figure-eight but also regenerates the transposon donor plasmid. To confirm the replicative mechanism, we have also used the Escherichia coli terminators (terC) which, when bound by the Tus protein, inhibit replication forks in a polarised manner. Finally, we demonstrate that the primase DnaG is essential, implicating a host-specific replication pathway.
Figures

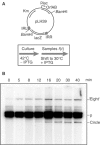
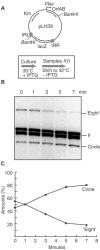
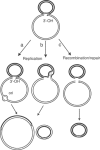
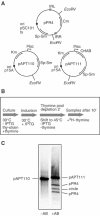



References
-
- Bussiere DE, Bastia D (1999) Termination of DNA replication of bacterial and plasmid chromosomes. Mol Microbiol 31: 1611–1618 - PubMed
-
- Chaconas G, Harshey RM (2002) Transposition of phage Mu DNA. In: Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), Washington, DC: ASM Press
-
- Chandler M, Mahillon J (2002) Insertion sequences revisited. In Mobile DNA II, Craig N, Craigie R, Gellert M, Lambowitz A (eds), pp 305–366. Washington, DC: ASM Press
-
- Churchward G, Belin D, Nagamine Y (1984) A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene 31: 165–171 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

