Ribosomal protein L7a binds RNA through two distinct RNA-binding domains
- PMID: 15361074
- PMCID: PMC1134697
- DOI: 10.1042/BJ20040371
Ribosomal protein L7a binds RNA through two distinct RNA-binding domains
Abstract
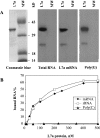
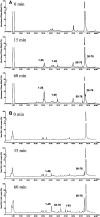
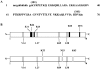
The human ribosomal protein L7a is a component of the major ribosomal subunit. We previously identified three nuclear-localization-competent domains within L7a, and demonstrated that the domain defined by aa (amino acids) 52-100 is necessary, although not sufficient, to target the L7a protein to the nucleoli. We now demonstrate that L7a interacts in vitro with a presumably G-rich RNA structure, which has yet to be defined. We also demonstrate that the L7a protein contains two RNA-binding domains: one encompassing aa 52-100 (RNAB1) and the other encompassing aa 101-161 (RNAB2). RNAB1 does not contain any known nucleic-acid-binding motif, and may thus represent a new class of such motifs. On the other hand, a specific region of RNAB2 is highly conserved in several other protein components of the ribonucleoprotein complex. We have investigated the topology of the L7a-RNA complex using a recombinant form of the protein domain that encompasses residues 101-161 and a 30mer poly(G) oligonucleotide. Limited proteolysis and cross-linking experiments, and mass spectral analyses of the recombinant protein domain and its complex with poly(G) revealed the RNA-binding region.
Figures






References
-
- Hernandez-Verdun D., Roussel P., Gebrane-Younes J. Emerging concepts of nucleolar assembly. J. Cell Sci. 2002;115:2265–2270. - PubMed
-
- Rudt F., Pieler T. Cytosolic import factor- and Ran-independent nuclear transport of ribosomal protein L5. Eur. J. Cell Biol. 2001;80:661–668. - PubMed
-
- Russo G., Ricciardelli G., Pietropaolo C. Different domains cooperate to target the human ribosomal L7a protein to the nucleus and to the nucleoli. J. Biol. Chem. 1997;272:5229–5235. - PubMed
-
- Damelin M., Silver P. A., Corbett A. H. Nuclear protein transport. Methods Enzymol. 2002;351:587–607. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

