Domain swapping is a consequence of minimal frustration
- PMID: 15361578
- PMCID: PMC518834
- DOI: 10.1073/pnas.0403724101
Domain swapping is a consequence of minimal frustration
Abstract
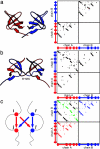
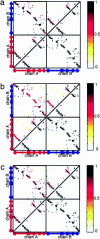
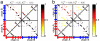
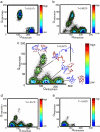
The same energy landscape principles associated with the folding of proteins into their monomeric conformations should also describe how these proteins oligomerize into domain-swapped conformations. We tested this hypothesis by using a simplified model for the epidermal growth factor receptor pathway substrate 8 src homology 3 domain protein, both of whose monomeric and domain-swapped structures have been solved. The model, which we call the symmetrized Gō-type model, incorporates only information regarding the monomeric conformation in an energy function for the dimer to predict the domain-swapped conformation. A striking preference for the correct domain-swapped structure was observed, indicating that overall monomer topology is a main determinant of the structure of domain-swapped dimers. Furthermore, we explore the free energy surface for domain swapping by using our model to characterize the mechanism of oligomerization.
Figures

References
-
- Bryngelson, J. D., Onuchic, J. N., Socci, N. D. & Wolynes, P. G. (1995) Proteins Struct. Funct. Genet. 21, 167–195. - PubMed
-
- Onuchic, J. N., Luthey-Schulten, Z. & Wolynes, P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600. - PubMed
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

