Continued colonization of the human genome by mitochondrial DNA
- PMID: 15361937
- PMCID: PMC515365
- DOI: 10.1371/journal.pbio.0020273
Continued colonization of the human genome by mitochondrial DNA
Abstract
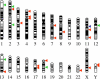
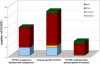
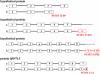
Integration of mitochondrial DNA fragments into nuclear chromosomes (giving rise to nuclear DNA sequences of mitochondrial origin, or NUMTs) is an ongoing process that shapes nuclear genomes. In yeast this process depends on double-strand-break repair. Since NUMTs lack amplification and specific integration mechanisms, they represent the prototype of exogenous insertions in the nucleus. From sequence analysis of the genome of Homo sapiens, followed by sampling humans from different ethnic backgrounds, and chimpanzees, we have identified 27 NUMTs that are specific to humans and must have colonized human chromosomes in the last 4-6 million years. Thus, we measured the fixation rate of NUMTs in the human genome. Six such NUMTs show insertion polymorphism and provide a useful set of DNA markers for human population genetics. We also found that during recent human evolution, Chromosomes 18 and Y have been more susceptible to colonization by NUMTs. Surprisingly, 23 out of 27 human-specific NUMTs are inserted in known or predicted genes, mainly in introns. Some individuals carry a NUMT insertion in a tumor-suppressor gene and in a putative angiogenesis inhibitor. Therefore in humans, but not in yeast, NUMT integrations preferentially target coding or regulatory sequences. This is indeed the case for novel insertions associated with human diseases and those driven by environmental insults. We thus propose a mutagenic phenomenon that may be responsible for a variety of genetic diseases in humans and suggest that genetic or environmental factors that increase the frequency of chromosome breaks provide the impetus for the continued colonization of the human genome by mitochondrial DNA.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures



References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. - PubMed
-
- Bensasson D, Feldman MW, Petrov DA. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 2003;57:343–354. - PubMed
-
- Blanchard JL, Schmidt GW. Mitochondrial DNA migration events in yeast and humans: Integration by a common end-joining mechanism and alternative perspectives on nucleotide substitution patterns. Mol Biol Evol. 1996;13:893. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

