A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs
- PMID: 15362974
- PMCID: PMC1134123
- DOI: 10.1042/BJ20041498
A novel UBA and UBX domain protein that binds polyubiquitin and VCP and is a substrate for SAPKs
Abstract
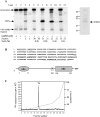
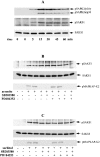
A widely expressed protein containing UBA (ubiquitin-associated) and UBX (ubiquitin-like) domains was identified as a substrate of SAPKs (stress-activated protein kinases). Termed SAKS1 (SAPK substrate-1), it was phosphorylated efficiently at Ser200 in vitro by SAPK3/p38gamma, SAPK4/p38delta and JNK (c-Jun N-terminal kinase), but weakly by SAPK2a/p38alpha, SAPK2b/p38beta2 or ERK (extracellular-signal-regulated kinase) 2. Ser200, situated immediately N-terminal to the UBX domain, became phosphorylated in HEK-293 (human embryonic kidney) cells in response to stressors. Phosphorylation was not prevented by SB 203580 (an inhibitor of SAPK2a/p38alpha and SAPK2b/p38beta2) and/or PD 184352 (which inhibits the activation of ERK1 and ERK2), and was similar in fibroblasts lacking both SAPK3/p38gamma and SAPK4/p38delta or JNK1 and JNK2. SAKS1 bound ubiquitin tetramers and VCP (valosin-containing protein) in vitro via the UBA and UBX domains respectively. The amount of VCP in cell extracts that bound to immobilized GST (glutathione S-transferase)-SAKS1 was enhanced by elevating the level of polyubiquitinated proteins, while SAKS1 and VCP in extracts were coimmunoprecipitated with an antibody raised against S5a, a component of the 19 S proteasomal subunit that binds polyubiquitinated proteins. PNGase (peptide N-glycanase) formed a 1:1 complex with VCP and, for this reason, also bound to immobilized GST-SAKS1. We suggest that SAKS1 may be an adaptor that directs VCP to polyubiquitinated proteins, and PNGase to misfolded glycoproteins, facilitating their destruction by the proteasome.
Figures




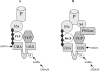
References
-
- McLaughlin M. M., Kumar S., McDonnell P. C., Van Horn S., Lee J. C., Livi G. P., Young P. R. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem. 1996;271:8488–8492. - PubMed
-
- Balendran A., Casamayor A., Deak M., Paterson A., Gaffney P., Currie R., Downes C. P., Alessi D. R. PDK1 acquires PDK2 activity in the presence of a synthetic peptide derived from the carboxyl terminus of PRK2. Curr. Biol. 1999;22:393–404. - PubMed
-
- Yaffe M. B., Lebarc G. G., Lai J., Obata T., Volinia S., Cantley L. C. A motif-based profile scanning approach for genome-wide prediction of signaling pathways. Nat. Biotechnol. 2001;19:348–353. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

