An integrated 4249 marker FISH/RH map of the canine genome
- PMID: 15363096
- PMCID: PMC520820
- DOI: 10.1186/1471-2164-5-65
An integrated 4249 marker FISH/RH map of the canine genome
Abstract
Background: The 156 breeds of dog recognized by the American Kennel Club offer a unique opportunity to map genes important in genetic variation. Each breed features a defining constellation of morphological and behavioral traits, often generated by deliberate crossing of closely related individuals, leading to a high rate of genetic disease in many breeds. Understanding the genetic basis of both phenotypic variation and disease susceptibility in the dog provides new ways in which to dissect the genetics of human health and biology.
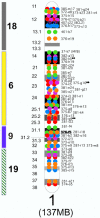
Results: To facilitate both genetic mapping and cloning efforts, we have constructed an integrated canine genome map that is both dense and accurate. The resulting resource encompasses 4249 markers, and was constructed using the RHDF5000-2 whole genome radiation hybrid panel. The radiation hybrid (RH) map features a density of one marker every 900 Kb and contains 1760 bacterial artificial chromosome clones (BACs) localized to 1423 unique positions, 851 of which have also been mapped by fluorescence in situ hybridization (FISH). The two data sets show excellent concordance. Excluding the Y chromosome, the map features an RH/FISH mapped BAC every 3.5 Mb and an RH mapped BAC-end, on average, every 2 Mb. For 2233 markers, the orthologous human genes have been established, allowing the identification of 79 conserved segments (CS) between the dog and human genomes, dramatically extending the length of most previously described CS.
Conclusions: These results provide a necessary resource for the canine genome mapping community to undertake positional cloning experiments and provide new insights into the comparative canine-human genome maps.
Figures

References
-
- Sargan DR, Yang F, Squire M, Milne BS, O'Brien PC, Ferguson-Smith MA. Use of flow-sorted canine chromosomes in the assignment of canine linkage, radiation hybrid, and syntenic groups to chromosomes: refinement and verification of the comparative chromosome map for dog and human. Genomics. 2000;69:182–95. doi: 10.1006/geno.2000.6334. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

