Identification of mammalian microRNA host genes and transcription units
- PMID: 15364901
- PMCID: PMC524413
- DOI: 10.1101/gr.2722704
Identification of mammalian microRNA host genes and transcription units
Abstract
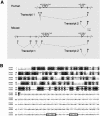
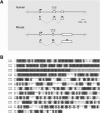
To derive a global perspective on the transcription of microRNAs (miRNAs) in mammals, we annotated the genomic position and context of this class of noncoding RNAs (ncRNAs) in the human and mouse genomes. Of the 232 known mammalian miRNAs, we found that 161 overlap with 123 defined transcription units (TUs). We identified miRNAs within introns of 90 protein-coding genes with a broad spectrum of molecular functions, and in both introns and exons of 66 mRNA-like noncoding RNAs (mlncRNAs). In addition, novel families of miRNAs based on host gene identity were identified. The transcription patterns of all miRNA host genes were curated from a variety of sources illustrating spatial, temporal, and physiological regulation of miRNA expression. These findings strongly suggest that miRNAs are transcribed in parallel with their host transcripts, and that the two different transcription classes of miRNAs ('exonic' and 'intronic') identified here may require slightly different mechanisms of biogenesis.
Figures
References
-
- Al-Shahrour, F., Diaz-Uriarte, R., and Dopazo, J. 2004. FatiGO: A web tool for finding significant associations of gene ontology terms with groups of genes. Bioinformatics 20: 578-580. - PubMed
-
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10: 428-433. - PubMed
-
- Bartel, D.P. 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281-297. - PubMed
-
- Bejerano, G., Pheasant, M., Makunin, I., Stephen, S., Kent, W.J., Mattick, J.S., and Haussler, D. 2004. Ultraconserved elements in the human genome. Science 304: 1321-1325. - PubMed
WEB SITE REFERENCES
-
- http://biobases.ibch.poznan.pl/ncRNA/; Noncoding RNAs database.
-
- http://www.bioinfo.rpi.edu/applications/mfold/; mfold RNA program.
-
- http://www.ensembl.org; ENSEMBL genome viewer.
-
- http://expression.gnf.org/cgi-bin/index.cgi; Gene expression atlas database.
-
- http://fantom2.gsc.riken.go.jp/; FANTOM functional annotation database.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
