Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis
- PMID: 15364960
- PMCID: PMC2172102
- DOI: 10.1083/jcb.200404158
Mathematical modeling reveals threshold mechanism in CD95-induced apoptosis
Abstract
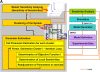
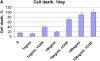
Mathematical modeling is required for understanding the complex behavior of large signal transduction networks. Previous attempts to model signal transduction pathways were often limited to small systems or based on qualitative data only. Here, we developed a mathematical modeling framework for understanding the complex signaling behavior of CD95(APO-1/Fas)-mediated apoptosis. Defects in the regulation of apoptosis result in serious diseases such as cancer, autoimmunity, and neurodegeneration. During the last decade many of the molecular mechanisms of apoptosis signaling have been examined and elucidated. A systemic understanding of apoptosis is, however, still missing. To address the complexity of apoptotic signaling we subdivided this system into subsystems of different information qualities. A new approach for sensitivity analysis within the mathematical model was key for the identification of critical system parameters and two essential system properties: modularity and robustness. Our model describes the regulation of apoptosis on a systems level and resolves the important question of a threshold mechanism for the regulation of apoptosis.
Figures











References
-
- Alon, U., M.G. Surette, N. Barkai, and S. Leibler. 1999. Robustness in bacterial chemotaxis. Nature. 397:168–171. - PubMed
-
- Ashkenazi, A., and V. Dixit. 1999. Apoptosis control by death and decoy receptors. Curr. Opin. Cell Biol. 11:255–260. - PubMed
-
- Bentele, M., and R. Eils. 2004. General stochastic hybrid method for the simulation of chemical reaction processes in cells. In CMSB ‘04. Lecture Notes in Computer Science. Springer, Heidelberg. In press.
-
- Bhalla, U.S., and R. Iyengar. 1999. Emergent properties of networks of biological signaling pathways. Science. 283:381–387. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

