Epidemiology and molecular characterization of Streptococcus pyogenes recovered from scarlet fever patients in central Taiwan from 1996 to 1999
- PMID: 15364982
- PMCID: PMC516299
- DOI: 10.1128/JCM.42.9.3998-4006.2004
Epidemiology and molecular characterization of Streptococcus pyogenes recovered from scarlet fever patients in central Taiwan from 1996 to 1999
Abstract
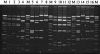
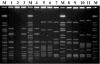
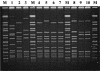
One hundred seventy-nine Streptococcus pyogenes isolates recovered from scarlet fever patients from 1996 to 1999 in central Taiwan were characterized by emm, Vir, and pulsed-field gel electrophoresis (PFGE) typing methods. The protocols for Vir and PFGE typing were standardized. A database of the DNA fingerprints for the isolates was established. Nine emm or emm-like genes, 19 Vir patterns, and 26 SmaI PFGE patterns were detected among the isolates. Among the three typing methods, PFGE was the most discriminatory. However, it could not completely replace Vir typing because some isolates with identical PFGE patterns could be further differentiated into several Vir patterns. The prevalent emm types were emm4 (n = 81 isolates [45%]), emm12 (n = 64 [36%]), emm1 (n = 14 [8%]), and emm22 (n = 13 [7%]). Some emm type isolates could be further differentiated into several emm-Vir-PFGE genotypes; however, only one genotype in each emm group was usually predominant. DNA from nine isolates was resistant to SmaI digestion. Further PFGE analysis with SgrAI showed that the SmaI digestion-resistant strains could be derived from indigenous strains by horizontal transfer of exogenous genetic material. The emergence of the new strains could have resulted in an increase in scarlet fever cases in central Taiwan since 2000. The emm sequences, Vir, and PFGE pattern database will serve as a basis for information for the long-term evolutionary study of local S. pyogenes strains.
Figures




Similar articles
-
Association of the shuffling of Streptococcus pyogenes clones and the fluctuation of scarlet fever cases between 2000 and 2006 in central Taiwan.BMC Microbiol. 2009 Jun 1;9:115. doi: 10.1186/1471-2180-9-115. BMC Microbiol. 2009. PMID: 19486515 Free PMC article.
-
Molecular epidemiology of group A streptococcus causing scarlet fever in northern Taiwan, 2001-2002.Diagn Microbiol Infect Dis. 2007 Jul;58(3):289-95. doi: 10.1016/j.diagmicrobio.2007.01.013. Epub 2007 May 29. Diagn Microbiol Infect Dis. 2007. PMID: 17532590
-
Molecular characterization of Group A streptococcal isolates causing scarlet fever and pharyngitis among young children: a retrospective study from a northern Taiwan medical center.J Microbiol Immunol Infect. 2014 Aug;47(4):304-10. doi: 10.1016/j.jmii.2013.02.007. Epub 2013 Apr 30. J Microbiol Immunol Infect. 2014. PMID: 23639381
-
Clonal differences among erythromycin-resistant Streptococcus pyogenes in Spain.Emerg Infect Dis. 1999 Mar-Apr;5(2):235-40. doi: 10.3201/eid0502.990207. Emerg Infect Dis. 1999. PMID: 10221875 Free PMC article. Review.
-
Prevalence and persistence of certain serologic types of Streptococcus pyogenes in metropolitan Tokyo.J Infect Chemother. 2006 Apr;12(2):53-62. doi: 10.1007/s10156-006-0432-4. J Infect Chemother. 2006. PMID: 16648943 Review.
Cited by
-
Genetic analysis of group A streptococcus isolates recovered during acute glomerulonephritis outbreaks in Guizhou Province of China.J Clin Microbiol. 2009 Mar;47(3):715-20. doi: 10.1128/JCM.00747-08. Epub 2008 Dec 30. J Clin Microbiol. 2009. PMID: 19116348 Free PMC article.
-
Clustered Regularly Interspaced Short Palindromic Repeats Are emm Type-Specific in Highly Prevalent Group A Streptococci.PLoS One. 2015 Dec 28;10(12):e0145223. doi: 10.1371/journal.pone.0145223. eCollection 2015. PLoS One. 2015. PMID: 26710228 Free PMC article.
-
Association of the shuffling of Streptococcus pyogenes clones and the fluctuation of scarlet fever cases between 2000 and 2006 in central Taiwan.BMC Microbiol. 2009 Jun 1;9:115. doi: 10.1186/1471-2180-9-115. BMC Microbiol. 2009. PMID: 19486515 Free PMC article.
-
A mini review of the pathogenesis of acute rheumatic fever and rheumatic heart disease.Front Cell Infect Microbiol. 2025 Apr 10;15:1447149. doi: 10.3389/fcimb.2025.1447149. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40276383 Free PMC article. Review.
-
The rise and fall of acute rheumatic fever and rheumatic heart disease: a mini review.Front Cardiovasc Med. 2023 May 23;10:1183606. doi: 10.3389/fcvm.2023.1183606. eCollection 2023. Front Cardiovasc Med. 2023. PMID: 37288267 Free PMC article. Review.
References
-
- Bisno, A. L., M. O. Brito, and C. M. Collins. 2003. Molecular basis of group A streptococcal virulence. Lancet Infect. Dis. 3:191-200. - PubMed
-
- Centers for Disease Control and Prevention. 2000. Multistate outbreak of listeriosis—United States, 2000. Morb. Mortal. Wkly. Rep. 49:1129-1130. - PubMed
-
- Chao, T. Y., H. C. Lu, C. S. Chiou, H. C. Chen, T. M. Pan, and K. T. Chen. 1998. An epidemiological investigation of a scarlet fever outbreak at a kindergarten in Taichung City. Epidemiol. Bull. 14:113-121.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

