Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR
- PMID: 15364984
- PMCID: PMC516350
- DOI: 10.1128/JCM.42.9.4016-4024.2004
Identification to the species level and differentiation between strains of Aspergillus clinical isolates by automated repetitive-sequence-based PCR
Abstract
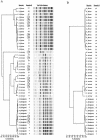
A commercially available repetitive-sequence-based PCR (rep-PCR) DNA fingerprinting assay adapted to an automated format, the DiversiLab system, enables rapid microbial identification and strain typing. We explored the performance of the DiversiLab system as a molecular typing tool for 69 Aspergillus isolates (38 A. fumigatus, 15 A. flavus, and 16 A. terreus isolates) had been previously characterized by morphological analysis. Initially, 27 Aspergillus isolates (10 A. fumigatus, 9 A. flavus, and 8 A. terreus isolates) were used as controls to create a rep-PCR-based DNA fingerprint library with the DiversiLab software. Then, 42 blinded Aspergillus isolates were typed using the system. The rep-PCR-based profile revealed 98% concordance with morphology-based identification. rep-PCR-based DNA fingerprints were reproducible and were consistent for DNA from both hyphae and conidia. DiversiLab dendrogram reports correctly identified all A. fumigatus (n = 28), A. terreus (n = 8), and A. flavus (n = 6) isolates in the 42 blinded Aspergillus isolates. rep-PCR-based identification of all isolates was 100% in agreement with the contiguous internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) sequence-based identification of the respective isolates. Additionally, the DiversiLab system could demonstrate strain-level differentiation of A. flavus and A. terreus. Automated rep-PCR may be a time-efficient, effective, easy-to-use, novel genotyping tool for identifying and determining the strain relatedness of fungi. This system may be useful for epidemiological studies, molecular typing, and surveillance of Aspergillus species.
Figures
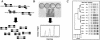



Similar articles
-
Species identification and strain differentiation of clinical Candida isolates using the DiversiLab system of automated repetitive sequence-based PCR.J Med Microbiol. 2007 Jun;56(Pt 6):778-787. doi: 10.1099/jmm.0.47106-0. J Med Microbiol. 2007. PMID: 17510263
-
Comparison of a semi-automated rep-PCR system and multilocus sequence typing for differentiation of Salmonella enterica isolates.J Microbiol Methods. 2010 Apr;81(1):11-6. doi: 10.1016/j.mimet.2010.01.013. Epub 2010 Jan 28. J Microbiol Methods. 2010. PMID: 20114063
-
Microbial DNA typing by automated repetitive-sequence-based PCR.J Clin Microbiol. 2005 Jan;43(1):199-207. doi: 10.1128/JCM.43.1.199-207.2005. J Clin Microbiol. 2005. PMID: 15634972 Free PMC article.
-
Molecular typing of Aspergillus species.Mycoses. 2008 Nov;51(6):463-76. doi: 10.1111/j.1439-0507.2008.01538.x. Epub 2008 Sep 12. Mycoses. 2008. PMID: 18793268 Review.
-
Aspergillus terreus complex.Med Mycol. 2009;47 Suppl 1:S42-6. doi: 10.1080/13693780802562092. Epub 2009 Mar 17. Med Mycol. 2009. PMID: 19291598 Review.
Cited by
-
Development of a DNA microarray for detection and identification of fungal pathogens involved in invasive mycoses.J Clin Microbiol. 2005 Oct;43(10):4943-53. doi: 10.1128/JCM.43.10.4943-4953.2005. J Clin Microbiol. 2005. PMID: 16207946 Free PMC article.
-
Aspergillus diversity in the environments of nosocomial infection cases at a university hospital.J Med Life. 2019 Apr-Jun;12(2):128-132. doi: 10.25122/jml-2018-0057. J Med Life. 2019. PMID: 31406513 Free PMC article.
-
Emerging Trichosporon asahii in elderly patients: epidemiological and molecular analysis by the DiversiLab system.Eur J Clin Microbiol Infect Dis. 2014 Sep;33(9):1497-503. doi: 10.1007/s10096-014-2099-6. Epub 2014 Apr 10. Eur J Clin Microbiol Infect Dis. 2014. PMID: 24718613
-
Development of RFLP-PCR method for the identification of medically important Aspergillus species using single restriction enzyme MwoI.Braz J Microbiol. 2014 Aug 29;45(2):503-7. doi: 10.1590/s1517-83822014000200018. eCollection 2014. Braz J Microbiol. 2014. PMID: 25242934 Free PMC article.
-
Clinical evaluation of the DiversiLab microbial typing system using repetitive-sequence-based PCR for characterization of Staphylococcus aureus strains.J Clin Microbiol. 2005 Mar;43(3):1187-92. doi: 10.1128/JCM.43.3.1187-1192.2005. J Clin Microbiol. 2005. PMID: 15750081 Free PMC article.
References
-
- Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. - PubMed
-
- Bart-Delabesse, E., C. Cordonnier, and S. Bretagne. 1999. Usefulness of genotyping with microsatellite markers to investigate hospital-acquired invasive aspergillosis. J. Hosp. Infect. 42:321-327. - PubMed
-
- Bart-Delabesse, E., J. Sarfati, J. P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J. P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. - PMC - PubMed
-
- Bertout, S., F. Renaud, T. De Meeus, M. A. Piens, B. Lebeau, M. A. Viviani, M. Mallie, J. M. Bastide, et al. 2000. Multilocus enzyme electrophoresis analysis of Aspergillus fumigatus strains isolated from the first clinical sample from patients with invasive aspergillosis. J. Med. Microbiol. 49:375-381. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

