Characterizations of adenovirus type 41 isolates from children with acute gastroenteritis in Japan, Vietnam, and Korea
- PMID: 15364986
- PMCID: PMC516313
- DOI: 10.1128/JCM.42.9.4032-4039.2004
Characterizations of adenovirus type 41 isolates from children with acute gastroenteritis in Japan, Vietnam, and Korea
Abstract
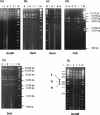
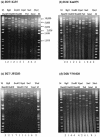
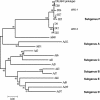
Genetic and antigenic characterizations of 70 strains of adenovirus type 41 (Ad41), isolated between 1998 and 2001 from children in Japan, Vietnam, and Korea, were done by DNA restriction enzyme (RE) analysis, sequencing analysis, and monoclonal antibody (MAb)-based enzyme-linked immunosorbent assay (ELISA). Eight genome types were observed in the present study, among which D25, D26, D27, and D28 were novel genome types. These eight genome types were divided into two genome-type clusters (GTCs) based on phylogenetic analysis of the hypervariable regions (HVRs) of the hexon. GTC1 includes D1, D25, D26, D27, and D28, and the GTC2 contains D4, D12, and D22. The amino acid homologies among the members within a GTC were 97 to 100%, whereas between the members of different GTCs the homologies were 92 to 94%. The specificity of the GTC classification was confirmed by ELISA with MAb 1F, which was selected by the Ad41 prototype Tak strain. It was found that only the isolates of GTC1 but not of GTC2 reacted with MAb 1F. These results suggest that Ad41 isolates from the three countries should be classified into two subtypes. The accumulation of amino acid mutations located in HVRs of hexon are indicative for the classification of Ad41 subtype.
Figures

Similar articles
-
Circulation of a novel pattern of infections by enteric adenovirus serotype 41 among children below 5 years of age in Kolkata, India.J Clin Microbiol. 2011 Feb;49(2):500-5. doi: 10.1128/JCM.01834-10. Epub 2010 Dec 1. J Clin Microbiol. 2011. PMID: 21123530 Free PMC article.
-
Prevalence and characterization of enteric adenoviruses in the South of Ireland.J Med Virol. 2007 Oct;79(10):1518-26. doi: 10.1002/jmv.20975. J Med Virol. 2007. PMID: 17705179
-
Molecular epidemiology of subgenus F adenoviruses associated with pediatric gastroenteritis during eight years in Hiroshima Prefecture as a limited area.Arch Virol. 2006 Dec;151(12):2511-7. doi: 10.1007/s00705-006-0816-x. Epub 2006 Jul 17. Arch Virol. 2006. PMID: 16847553
-
Molecular characterization of enteric adenovirus genotypes 40 and 41 identified in children with acute gastroenteritis in Kolkata, India during 2013-2014.J Med Virol. 2017 Apr;89(4):606-614. doi: 10.1002/jmv.24672. Epub 2016 Sep 19. J Med Virol. 2017. PMID: 27584661
-
An outbreak of adenovirus serotype 41 infection in infants and children with acute gastroenteritis in Maizuru City, Japan.Infect Genet Evol. 2007 Mar;7(2):279-84. doi: 10.1016/j.meegid.2006.11.005. Epub 2006 Dec 6. Infect Genet Evol. 2007. PMID: 17157081
Cited by
-
Molecular subtypes of Adenovirus-associated acute respiratory infection outbreak in children in Northern Vietnam and risk factors of more severe cases.PLoS Negl Trop Dis. 2023 Nov 7;17(11):e0011311. doi: 10.1371/journal.pntd.0011311. eCollection 2023 Nov. PLoS Negl Trop Dis. 2023. PMID: 37934746 Free PMC article.
-
Molecular and epidemiological characterization of human adenovirus and classic human astrovirus in children with acute diarrhea in Shanghai, 2017-2018.BMC Infect Dis. 2021 Jul 29;21(1):713. doi: 10.1186/s12879-021-06403-1. BMC Infect Dis. 2021. PMID: 34325664 Free PMC article.
-
Circulation of a novel pattern of infections by enteric adenovirus serotype 41 among children below 5 years of age in Kolkata, India.J Clin Microbiol. 2011 Feb;49(2):500-5. doi: 10.1128/JCM.01834-10. Epub 2010 Dec 1. J Clin Microbiol. 2011. PMID: 21123530 Free PMC article.
-
Molecular Epidemiology of Human Adenovirus from Acute Gastroenteritis Cases in Brazil After the COVID-19 Pandemic Period, 2021-2023.Viruses. 2025 Apr 17;17(4):577. doi: 10.3390/v17040577. Viruses. 2025. PMID: 40285019 Free PMC article.
-
Prevalence and molecular characterisation of human adenovirus in diarrhoeic children in Tanzania; a case control study.BMC Infect Dis. 2014 Dec 12;14:666. doi: 10.1186/s12879-014-0666-1. BMC Infect Dis. 2014. PMID: 25495029 Free PMC article.
References
-
- Adrian, T., B. Best, and R. Wigand. 1985. A proposal for naming adenovirus genome types, exemplified by adenovirus type 6. J. Gen. Virol. 66:2685-2691. - PubMed
-
- Akusjarvi, G., P. Alestrom, M. Pettersson, M. Lager, H. Jornvall, and U. Pettersson. 1984. The gene for the adenovirus 2 hexon polypeptide. J. Biol. Chem. 259:13976-13979. - PubMed
-
- de Jong, J. C., K. Bijlsma, A. G. Wermenbol, M. W. Verweij-Uijterwaal, H. G. van der Avoort, D. J. Wood, A. S. Bailey, and A. D. Osterhaus. 1993. Detection, typing, and subtyping of enteric adenoviruses 40 and 41 from fecal samples and observation of changing incidences of infections with these types and subtypes. J. Clin. Microbiol. 31:1562-1569. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

