Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India
- PMID: 15364997
- PMCID: PMC516302
- DOI: 10.1128/JCM.42.9.4111-4120.2004
Virulence characteristics and molecular epidemiology of enteroaggregative Escherichia coli isolates from hospitalized diarrheal patients in Kolkata, India
Abstract
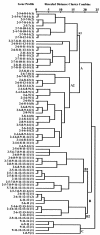
Enteroaggregative Escherichia coli (EAEC) is an important diarrheal enteropathogen defined by aggregative adherence to cultured epithelial cells. We have detected EAEC from 121 (6.6%) of 1,826 hospitalized patients admitted with diarrhea to the Infectious Diseases Hospital in Kolkata, India. Watery diarrhea was recorded significantly (P = 0.0142) more often in children. The majority of the EAEC isolates were not serotypeable (62%) and showed resistance to five or more antibiotics (76%). We studied different virulence genes and the molecular epidemiology of 121 EAEC isolates recovered from diarrheal patients. A PCR assay for detection of virulence genes, an assay for determination of clump formation in liquid culture, and a HeLa cell adherence assay were carried out to characterize the EAEC isolates. Investigations were also conducted to correlate the virulence gene profiles with diarrheal symptoms and molecular epidemiology by pulsed-field gel electrophoresis (PFGE). Two or more virulence genes were detected in 109 (90.1%) EAEC isolates. In the cluster analysis, some isolates with specific gene profiles and phenotypes formed a group or subcluster. This study highlights the comparative distributions of three fimbrial adhesins and other virulence genes among EAEC isolates. The diverse virulence gene and PFGE profiles, along with the existence of diverse serotypes and antibiograms, suggests that the EAEC isolates are genetically heterogeneous in Kolkata.
Figures


References
-
- Adachi, J. A., Z. D. Jiang, J. J. Mathewson, M. P. Verenkar, S. Thompson, F. Martinez-Sandoval, R. Steffen, C. D. Ericsson, and H. L. DuPont. 2001. Enteroaggregative Escherichia coli as a major etiologic agent in traveler's diarrhea in 3 regions of the world. Clin. Infect. Dis. 32:1706-1709. - PubMed
-
- Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. - PubMed
-
- Bhan, M. K., P. Raj, M. M. Levine, J. B. Kaper, N. Bhandari, R. Srivastava, R. Kumar, and S. Sazawal. 1989. Enteroaggregative Escherichia coli associated with persistent diarrhea in a cohort of rural children in India. J. Infect. Dis. 159:1061-1064. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

