Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X
- PMID: 15367597
- PMCID: PMC516385
- DOI: 10.1128/JVI.78.19.10310-10319.2004
Np9 protein of human endogenous retrovirus K interacts with ligand of numb protein X
Abstract
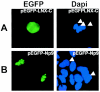
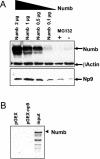
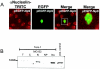
We have recently identified Np9 as a novel nuclear protein produced by the human endogenous retrovirus K and were able to document the exclusive presence of np9 transcript in tumors and transformed cells. With the aim of studying whether Np9 has a role in tumorigenesis, a systematic search for interacting proteins was performed. Here, we identify the RING-type E3 ubiquitin ligase LNX (ligand of Numb protein X) as an Np9-interacting partner. We furthermore show that the interaction involves N- and C-terminal domains of both proteins and can affect the subcellular localization of LNX. LNX has been reported to target the cell fate determinant and Notch antagonist Numb for proteasome-dependent degradation, thereby causing an increase in transactivational activity of Notch. We document that LNX-interacting Np9, like Numb, is unstable and degraded via the proteasome pathway and that ectopic Numb can stabilize recombinant Np9. Combined, these findings point to the possibility that Np9 affects tumorigenesis through the LNX/Numb/Notch pathway.
Figures







References
-
- Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800-1807. - PubMed
-
- Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinhard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. - PubMed
-
- Beverly, L. J., and A. J. Capobianco. 2003. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3:551-564. - PubMed
-
- Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

