Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth
- PMID: 15367602
- PMCID: PMC516403
- DOI: 10.1128/JVI.78.19.10360-10369.2004
Human cytomegalovirus UL84 insertion mutant defective for viral DNA synthesis and growth
Abstract
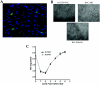
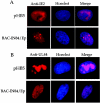
Human cytomegalovirus (HCMV) UL84 is required for oriLyt-dependent DNA replication, and evidence from transient transfection assays suggests that UL84 directly participates in DNA synthesis. In addition, because of its apparent interaction with IE2, UL84 is implicated as a possible regulatory protein. To address the role of UL84 in the context of the viral genome, we generated a recombinant HCMV bacterial artificial chromosome (BAC) construct that did not express the UL84 gene product. This construct, BAC-IN84/Ep, displayed a null phenotype in that it failed to produce infectious virus after transfection into human fibroblast cells, whereas a revertant virus readily produced viral plaques and, subsequently, infectious virus. Real-time quantitative PCR showed that BAC-IN84/Ep was defective for DNA synthesis in that no increase in the accumulation of viral DNA was observed in transfected cells. We were unable to complement BAC-IN84/Ep in trans; however, oriLyt-dependent DNA replication was observed by the cotransfection of UL84 and BAC-IN84/Ep. An analysis of viral mRNA by real-time PCR indicated that, even in the absence of DNA synthesis, all representative kinetic classes of genes were expressed in cells transfected with BAC-IN84/Ep. The detection of UL44 and IE2 by immunofluorescence in BAC-IN84/Ep-transfected cells showed that these proteins failed to partition into replication compartments, indicating that UL84 expression is essential for the formation of these proteins into replication centers within the context of the viral genome. These results show that UL84 provides an essential DNA replication function and influences the subcellular localization of other viral proteins.
Figures


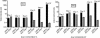
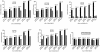
Similar articles
-
Human cytomegalovirus UL84 oligomerization and heterodimerization domains act as transdominant inhibitors of oriLyt-dependent DNA replication: evidence that IE2-UL84 and UL84-UL84 interactions are required for lytic DNA replication.J Virol. 2004 Sep;78(17):9203-14. doi: 10.1128/JVI.78.17.9203-9214.2004. J Virol. 2004. PMID: 15308715 Free PMC article.
-
Human cytomegalovirus UL84 localizes to the cell nucleus via a nuclear localization signal and is a component of viral replication compartments.J Virol. 2002 Sep;76(17):8931-8. doi: 10.1128/jvi.76.17.8931-8938.2002. J Virol. 2002. PMID: 12163612 Free PMC article.
-
Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays.J Virol. 1996 Nov;70(11):7398-413. doi: 10.1128/JVI.70.11.7398-7413.1996. J Virol. 1996. PMID: 8892858 Free PMC article.
-
Nuts and bolts of human cytomegalovirus lytic DNA replication.Curr Top Microbiol Immunol. 2008;325:153-66. doi: 10.1007/978-3-540-77349-8_9. Curr Top Microbiol Immunol. 2008. PMID: 18637505 Review.
-
Recombinant cytomegaloviruses for study of replication and pathogenesis.Intervirology. 1996;39(5-6):320-30. doi: 10.1159/000150503. Intervirology. 1996. PMID: 9130042 Review.
Cited by
-
The carboxy-terminal segment of the human cytomegalovirus DNA polymerase accessory subunit UL44 is crucial for viral replication.J Virol. 2010 Nov;84(21):11563-8. doi: 10.1128/JVI.01033-10. Epub 2010 Aug 25. J Virol. 2010. PMID: 20739543 Free PMC article.
-
Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus.J Virol. 2010 Sep;84(17):8409-21. doi: 10.1128/JVI.00189-10. Epub 2010 Jun 10. J Virol. 2010. PMID: 20538862 Free PMC article.
-
Nucleocytoplasmic shuttling of human cytomegalovirus UL84 is essential for virus growth.J Virol. 2010 Sep;84(17):8484-94. doi: 10.1128/JVI.00738-10. Epub 2010 Jun 23. J Virol. 2010. PMID: 20573826 Free PMC article.
-
Internal deletions of IE2 86 and loss of the late IE2 60 and IE2 40 proteins encoded by human cytomegalovirus affect the levels of UL84 protein but not the amount of UL84 mRNA or the loading and distribution of the mRNA on polysomes.J Virol. 2008 Nov;82(22):11383-97. doi: 10.1128/JVI.01293-08. Epub 2008 Sep 10. J Virol. 2008. PMID: 18787008 Free PMC article.
-
Strain-Dependent Restriction of Human Cytomegalovirus by Zinc Finger Antiviral Proteins.J Virol. 2023 Mar 30;97(3):e0184622. doi: 10.1128/jvi.01846-22. Epub 2023 Mar 14. J Virol. 2023. PMID: 36916924 Free PMC article.
References
-
- Abbate, J., J. C. Lacayo, M. Prichard, G. Pari, and M. A. McVoy. 2001. Bifunctional protein conferring enhanced green fluorescence and puromycin resistance. BioTechniques 31:336-340. - PubMed
-
- Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol 18:4899-4913. - PMC - PubMed
-
- Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

