RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration
- PMID: 15367605
- PMCID: PMC516422
- DOI: 10.1128/JVI.78.19.10390-10398.2004
RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration
Abstract
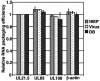
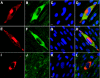
The assembly of human cytomegalovirus (HCMV) virions is a complex process and involves the incorporation of viral transcripts. These RNAs are delivered to the newly infected cells and have the potential to be translated in the absence of HCMV gene expression. We have quantified the relative amount of RNAs in HCMV virions and infected cells with real-time reverse transcription-PCR and observed that viral and cellular RNAs are packaged in proportion to the amount of RNA within the cell at the time of assembly. To determine whether cis elements influenced RNA packaging, we constructed a recombinant HCMV mutant virus that expressed the yellow fluorescence protein (YFP) gene fused to the virion RNA UL21.5. We also constructed a mutant virus in which the UL21.5 transcription unit was replaced with the YFP gene. YFP RNA was incorporated into both viruses, indicating that RNA is incorporated in the absence of a virus-specific signal motif. Furthermore, with in situ hybridization, packaged transcripts were observed throughout the cytoplasm of the infected cells, including the site of virus assembly. Several proteins that nonspecifically interact with RNA, including the tegument protein pp28, were found within HCMV virions. These studies demonstrate that both viral and cellular RNAs are nonspecifically incorporated into HCMV, potentially through interactions with several virion proteins.
Figures

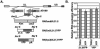

Similar articles
-
RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions.J Virol. 2001 Sep;75(17):8105-16. doi: 10.1128/jvi.75.17.8105-8116.2001. J Virol. 2001. PMID: 11483756 Free PMC article.
-
The tegument protein pp65 of human cytomegalovirus acts as an optional scaffold protein that optimizes protein uploading into viral particles.J Virol. 2014 Sep 1;88(17):9633-46. doi: 10.1128/JVI.01415-14. Epub 2014 Jun 11. J Virol. 2014. PMID: 24920816 Free PMC article.
-
Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex.J Virol. 2014 Aug;88(16):9086-99. doi: 10.1128/JVI.01141-14. Epub 2014 Jun 4. J Virol. 2014. PMID: 24899189 Free PMC article.
-
The importance of virion-incorporated cellular RNA-Binding Proteins in viral particle assembly and infectivity.Semin Cell Dev Biol. 2021 Mar;111:108-118. doi: 10.1016/j.semcdb.2020.08.002. Epub 2020 Sep 10. Semin Cell Dev Biol. 2021. PMID: 32921578 Free PMC article. Review.
-
Human cytomegalovirus tegument proteins (pp65, pp71, pp150, pp28).Virol J. 2012 Jan 17;9:22. doi: 10.1186/1743-422X-9-22. Virol J. 2012. PMID: 22251420 Free PMC article. Review.
Cited by
-
Human Cytomegalovirus pUL47 Modulates Tegumentation and Capsid Accumulation at the Viral Assembly Complex.J Virol. 2015 Jul;89(14):7314-28. doi: 10.1128/JVI.00603-15. Epub 2015 May 6. J Virol. 2015. PMID: 25948747 Free PMC article.
-
RNAs in the virion of Kaposi's sarcoma-associated herpesvirus.J Virol. 2005 Aug;79(16):10138-46. doi: 10.1128/JVI.79.16.10138-10146.2005. J Virol. 2005. PMID: 16051806 Free PMC article.
-
Adventitious Virus Detection in Cells by High-Throughput Sequencing of Newly Synthesized RNAs: Unambiguous Differentiation of Cell Infection from Carryover of Viral Nucleic Acids.mSphere. 2019 Jun 5;4(3):e00298-19. doi: 10.1128/mSphere.00298-19. mSphere. 2019. PMID: 31167947 Free PMC article.
-
Dynamic Epstein-Barr virus gene expression on the path to B-cell transformation.Adv Virus Res. 2014;88:279-313. doi: 10.1016/B978-0-12-800098-4.00006-4. Adv Virus Res. 2014. PMID: 24373315 Free PMC article. Review.
-
Estrogen-related receptor α is required for efficient human cytomegalovirus replication.Proc Natl Acad Sci U S A. 2014 Dec 30;111(52):E5706-15. doi: 10.1073/pnas.1422361112. Epub 2014 Dec 15. Proc Natl Acad Sci U S A. 2014. PMID: 25512541 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

