Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme
- PMID: 15367637
- PMCID: PMC516392
- DOI: 10.1128/JVI.78.19.10706-10714.2004
Forced selection of a human immunodeficiency virus type 1 variant that uses a non-self tRNA primer for reverse transcription: involvement of viral RNA sequences and the reverse transcriptase enzyme
Abstract
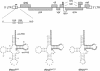
Human immunodeficiency virus type 1 uses the tRNA(3)(Lys) molecule as a selective primer for reverse transcription. This primer specificity is imposed by sequence complementarity between the tRNA primer and two motifs in the viral RNA genome: the primer-binding site (PBS) and the primer activation signal (PAS). In addition, there may be specific interactions between the tRNA primer and viral proteins, such as the reverse transcriptase (RT) enzyme. We constructed viruses with mutations in the PAS and PBS that were designed to employ the nonself primer tRNA(Pro) or tRNA(1,2)(Lys). These mutants exhibited a severe replication defect, indicating that additional adaptation of the mutant virus is required to accommodate the new tRNA primer. Multiple independent virus evolution experiments were performed to select for fast-replicating variants. Reversion to the wild-type PBS-lys3 sequence was the most frequent escape route. However, we identified one culture in which the virus gained replication capacity without reversion of the PBS. This revertant virus eventually optimized the PAS motif for interaction with the nonself primer. Interestingly, earlier evolution samples revealed a single amino acid change of an otherwise well-conserved residue in the RNase H domain of the RT enzyme, implicating this domain in selective primer usage. We demonstrate that both the PAS and RT mutations improve the replication capacity of the tRNA(1,2)(Lys)-using virus.
Figures


Similar articles
-
The G490E mutation in reverse transcriptase does not impact tRNA primer selection by HIV-1 with altered PBS and A-loop.Virology. 2006 Sep 1;352(2):380-9. doi: 10.1016/j.virol.2006.04.019. Epub 2006 Jun 15. Virology. 2006. PMID: 16781756
-
Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS.RNA. 2002 Mar;8(3):357-69. doi: 10.1017/s1355838202028194. RNA. 2002. PMID: 12003495 Free PMC article.
-
The tRNA primer activation signal in the human immunodeficiency virus type 1 genome is important for initiation and processive elongation of reverse transcription.J Virol. 2002 Mar;76(5):2329-39. doi: 10.1128/jvi.76.5.2329-2339.2002. J Virol. 2002. PMID: 11836411 Free PMC article.
-
tRNA(Lys3): the primer tRNA for reverse transcription in HIV-1.IUBMB Life. 2002 Feb;53(2):107-14. doi: 10.1080/15216540211469. IUBMB Life. 2002. PMID: 12049192 Review.
-
HIV-1 reverse transcription initiation: a potential target for novel antivirals?Virus Res. 2008 Jun;134(1-2):4-18. doi: 10.1016/j.virusres.2007.12.009. Epub 2008 Feb 6. Virus Res. 2008. PMID: 18255184 Review.
Cited by
-
Low-level alternative tRNA priming of reverse transcription of HIV-1 and SIV in vivo.Retrovirology. 2019 Apr 4;16(1):11. doi: 10.1186/s12977-019-0473-2. Retrovirology. 2019. PMID: 30947720 Free PMC article.
-
Hairpin-induced tRNA-mediated (HITME) recombination in HIV-1.Nucleic Acids Res. 2006 May 2;34(8):2206-18. doi: 10.1093/nar/gkl226. Print 2006. Nucleic Acids Res. 2006. PMID: 16670429 Free PMC article.
-
Heterogeneous structures formed by conserved RNA sequences within the HIV reverse transcription initiation site.RNA. 2016 Nov;22(11):1689-1698. doi: 10.1261/rna.056804.116. Epub 2016 Sep 9. RNA. 2016. PMID: 27613581 Free PMC article.
-
New insights into inhibition of human immunodeficiency virus type 1 replication through mutant tRNALys3.Retrovirology. 2013 Oct 24;10:112. doi: 10.1186/1742-4690-10-112. Retrovirology. 2013. PMID: 24156557 Free PMC article.
-
The interplay between virus infection and the cellular RNA interference machinery.FEBS Lett. 2006 May 22;580(12):2896-902. doi: 10.1016/j.febslet.2006.02.070. Epub 2006 Mar 6. FEBS Lett. 2006. PMID: 16563388 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

