Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses
- PMID: 15367642
- PMCID: PMC516408
- DOI: 10.1128/JVI.78.19.10755-10764.2004
Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses
Abstract
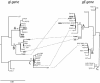
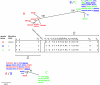
Herpes simplex virus type 1 (HSV-1) is a ubiquitous human pathogen which establishes lifelong infections. In the present study, we determined the sequence diversity of the complete genes coding for glycoproteins G (gG), I (gI), and E (gE), comprising 2.3% of the HSV-1 genome and located within the unique short (US) region, for 28 clinical HSV-1 isolates inducing oral lesions, genital lesions, or encephalitis. Laboratory strains F and KOS321 were sequenced in parallel. Phylogenetic analysis, including analysis of laboratory strain 17 (GenBank), revealed that the sequences were separated into three genetic groups. The identification of different genogroups facilitated the detection of recombinant viruses by using specific nucleotide substitutions as recombination markers. Seven of the isolates and strain 17 displayed sequences consistent with intergenic recombination, and at least four isolates were intragenic recombinants. The observed frequency of recombination based on an analysis of a short stretch of the US region suggests that most full-length HSV-1 genomes consist of a mosaic of segments from different genetic groups. Polymorphic tandem repeat regions, consisting of two to eight blocks of 21 nucleotides in the gI gene and seven to eight repeats of 3 nucleotides in the gG gene, were also detected. Laboratory strain KOS321 displayed a frameshift mutation in the gI gene with a subsequent alteration of the deduced intracellular portion of the protein. The presence of polymorphic tandem repeat regions and the different genogroup identities can be used for molecular epidemiology studies and for further detection of recombination in the HSV-1 genome.
Figures



Similar articles
-
Molecular characterization of clinical isolates of herpes simplex virus type 1 collected in a tertiary-care hospital in Dublin, Ireland.J Med Virol. 2013 May;85(5):839-44. doi: 10.1002/jmv.23541. J Med Virol. 2013. PMID: 23508909
-
Phylogenetic comparison of exonic US4, US7 and UL44 regions of clinical herpes simplex virus type 1 isolates showed lack of association between their anatomic sites of infection and genotypic/sub genotypic classification.Virol J. 2012 Mar 14;9:65. doi: 10.1186/1743-422X-9-65. Virol J. 2012. PMID: 22416856 Free PMC article.
-
A 10-year molecular survey of herpes simplex virus type 1 in Germany demonstrates a stable and high prevalence of genotypes A and B.J Clin Virol. 2009 Mar;44(3):235-7. doi: 10.1016/j.jcv.2008.12.016. Epub 2009 Jan 30. J Clin Virol. 2009. PMID: 19186100
-
History and genomic sequence analysis of the herpes simplex virus 1 KOS and KOS1.1 sub-strains.Virology. 2016 Jan;487:215-21. doi: 10.1016/j.virol.2015.09.026. Epub 2015 Nov 5. Virology. 2016. PMID: 26547038 Free PMC article. Review.
-
Evolution of herpes simplex virus type 1 under herpesviral evolutionary processes.Arch Virol. 1999;144(4):637-56. doi: 10.1007/s007050050533. Arch Virol. 1999. PMID: 10365158 Review.
Cited by
-
Ancient Recombination Events between Human Herpes Simplex Viruses.Mol Biol Evol. 2017 Jul 1;34(7):1713-1721. doi: 10.1093/molbev/msx113. Mol Biol Evol. 2017. PMID: 28369565 Free PMC article.
-
Shared ancestry of herpes simplex virus 1 strain Patton with recent clinical isolates from Asia and with strain KOS63.Virology. 2017 Dec;512:124-131. doi: 10.1016/j.virol.2017.09.016. Virology. 2017. PMID: 28957690 Free PMC article.
-
Evolution and diversity in human herpes simplex virus genomes.J Virol. 2014 Jan;88(2):1209-27. doi: 10.1128/JVI.01987-13. Epub 2013 Nov 13. J Virol. 2014. PMID: 24227835 Free PMC article.
-
Divergence and recombination of clinical herpes simplex virus type 2 isolates.J Virol. 2007 Dec;81(23):13158-67. doi: 10.1128/JVI.01310-07. Epub 2007 Sep 19. J Virol. 2007. PMID: 17881457 Free PMC article.
-
The Detection and Characterization of Herpes Simplex Virus Type 1 in Confirmed Measles Cases.Sci Rep. 2019 Sep 4;9(1):12785. doi: 10.1038/s41598-019-48994-5. Sci Rep. 2019. PMID: 31484944 Free PMC article. Clinical Trial.
References
-
- Baker, R. O., and J. D. Hall. 1998. Impaired mismatch extension by a herpes simplex DNA polymerase mutant with an editing nuclease defect. J. Biol. Chem. 273:24075-24082. - PubMed
-
- Bower, J. R., H. Mao, C. Durishin, E. Rozenbom, M. Detwiler, D. Rempinski, T. L. Karban, and K. S. Rosenthal. 1999. Intrastrain variants of herpes simplex virus type 1 isolated from a neonate with fatal disseminated infection differ in the ICP34.5 gene, glycoprotein processing, and neuroinvasiveness. J. Virol. 73:3843-3853. - PMC - PubMed
-
- Brown, S. M., J. H. Subak-Sharpe, J. Harland, and A. R. MacLean. 1992. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 73:293-301. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

