GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I
- PMID: 15367666
- PMCID: PMC516758
- DOI: 10.1128/MCB.24.19.8447-8456.2004
GRIM-19, a cell death regulatory protein, is essential for assembly and function of mitochondrial complex I
Abstract
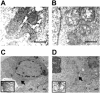
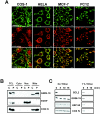
Mitochondria play essential roles in cellular energy production via the oxidative phosphorylation system (OXPHOS) consisting of five multiprotein complexes and also in the initiation of apoptosis. NADH:ubiquinone oxidoreductase (complex I) is the largest complex that catalyzes the first step of electron transfer in the OXPHOS system. GRIM-19 was originally identified as a nuclear protein with apoptotic nature in interferon (IFN)- and all-trans-retinoic acid (RA)-induced tumor cells. To reveal its biological role, we generated mice deficient in GRIM-19 by gene targeting. Homologous deletion of GRIM-19 causes embryonic lethality at embryonic day 9.5. GRIM-19(-/-) blastocysts show retarded growth in vitro and, strikingly, display abnormal mitochondrial structure, morphology, and cellular distribution. We reexamined the cellular localization of GRIM-19 in various cell types and found its primary localization in the mitochondria. Furthermore, GRIM-19 is detected in the native form of mitochondrial complex I. Finally, we show that elimination of GRIM-19 destroys the assembly and electron transfer activity of complex I and also influences the other complexes in the mitochondrial respiratory chain. Our result demonstrates that GRIM-19, a gene product with a specific role in IFN-RA-induced cell death, is a functional component of mitochondrial complex I and is essential for early embryonic development.
Figures





Similar articles
-
Coupling mitochondrial respiratory chain to cell death: an essential role of mitochondrial complex I in the interferon-beta and retinoic acid-induced cancer cell death.Cell Death Differ. 2007 Feb;14(2):327-37. doi: 10.1038/sj.cdd.4402004. Epub 2006 Jul 7. Cell Death Differ. 2007. PMID: 16826196
-
GRIM-19 in Health and Disease.Adv Anat Pathol. 2008 Jan;15(1):46-53. doi: 10.1097/PAP.0b013e31815e5258. Adv Anat Pathol. 2008. PMID: 18156812
-
GRIM-19 is essential for maintenance of mitochondrial membrane potential.Mol Biol Cell. 2008 May;19(5):1893-902. doi: 10.1091/mbc.e07-07-0683. Epub 2008 Feb 20. Mol Biol Cell. 2008. PMID: 18287540 Free PMC article.
-
Respiratory chain complex I deficiency.Am J Med Genet. 2001 Spring;106(1):37-45. doi: 10.1002/ajmg.1397. Am J Med Genet. 2001. PMID: 11579423 Review.
-
Complex I and Parkinson's disease.IUBMB Life. 2001 Sep-Nov;52(3-5):135-41. doi: 10.1080/15216540152845939. IUBMB Life. 2001. PMID: 11798025 Review.
Cited by
-
Thyroid Hürthle cell tumors: research of potential markers of malignancy.J Endocrinol Invest. 2016 Feb;39(2):153-8. doi: 10.1007/s40618-015-0356-x. Epub 2015 Jul 19. J Endocrinol Invest. 2016. PMID: 26188382
-
GRIM-19 and p16(INK4a) synergistically regulate cell cycle progression and E2F1-responsive gene expression.J Biol Chem. 2010 Sep 3;285(36):27545-52. doi: 10.1074/jbc.M110.105767. Epub 2010 Jun 3. J Biol Chem. 2010. PMID: 20522552 Free PMC article.
-
Neurospora strains harboring mitochondrial disease-associated mutations in iron-sulfur subunits of complex I.Genetics. 2005 Sep;171(1):91-9. doi: 10.1534/genetics.105.041517. Epub 2005 Jun 14. Genetics. 2005. PMID: 15956670 Free PMC article.
-
Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells.Stem Cell Res. 2015 Nov;15(3):678-693. doi: 10.1016/j.scr.2015.10.015. Epub 2015 Oct 31. Stem Cell Res. 2015. PMID: 26561938 Free PMC article.
-
Maternal enzyme masks the phenotype of mouse embryos lacking dihydrolipoamide dehydrogenase.Reprod Biomed Online. 2009 Jul;19(1):79-88. doi: 10.1016/s1472-6483(10)60050-8. Reprod Biomed Online. 2009. PMID: 19573295 Free PMC article.
References
-
- Akira, S., Y. Nishio, M. Inoue, X. J. Wang, S. Wei, T. Matsusaka, K. Yoshida, T. Sudo, M. Naruto, and T. Kishimoto. 1994. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77:63-71. - PubMed
-
- Angell, J. E., D. J. Lindner, P. S. Shapiro, E. R. Hofmann, and D. V. Kalvakolanu. 2000. Identification of GRIM-19, a novel cell death-regulatory gene induced by the interferon-beta and retinoic acid combination, using a genetic approach. J. Biol. Chem. 275:33416-33426. - PubMed
-
- Antonicka, H., I. Ogilvie, T. Taivassalo, R. P. Anitori, R. G. Haller, J. Vissing, N. G. Kennaway, and E. A. Shoubridge. 2003. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. J. Biol. Chem. 278:43081-43088. - PubMed
-
- Carroll, J., I. M. Fearnley, R. J. Shannon, J. Hirst, and J. E. Walker. 2003. Analysis of the subunit composition of complex I from bovine heart mitochondria. Mol. Cell. Proteomics 2:117-126. - PubMed
-
- Dahl, H. H., and D. R. Thorburn. 2001. Mitochondrial diseases: beyond the magic circle. Am. J. Med. Genet. 106:1-3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
