The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase
- PMID: 15367669
- PMCID: PMC516753
- DOI: 10.1128/MCB.24.19.8477-8486.2004
The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase
Abstract
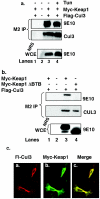
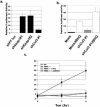
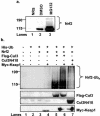
The Nrf2 transcription factor promotes survival following cellular insults that trigger oxidative damage. Nrf2 activity is opposed by the BTB/POZ domain protein Keap1. Keap1 is proposed to regulate Nrf2 activity strictly through its capacity to inhibit Nrf2 nuclear import. Recent work suggests that inhibition of Nrf2 may also depend upon ubiquitin-mediated proteolysis. To address the contribution of Keap1-dependent sequestration versus Nrf2 proteolysis, we identified the E3 ligase that regulates Nrf2 ubiquitination. We demonstrate that Keap1 is not solely a cytosolic anchor; rather, Keap1 is an adaptor that bridges Nrf2 to Cul3. We demonstrate that Cul3-Keap1 complexes regulate Nrf2 polyubiquitination both in vitro and in vivo. Inhibition of either Keap1 or Cul3 increases Nrf2 nuclear accumulation, leading to promiscuous activation of Nrf2-dependent gene expression. Our data demonstrate that Keap1 restrains Nrf2 activity via its capacity to target Nrf2 to a cytoplasmic Cul3-based E3 ligase and suggest a model in which Keap1 coordinately regulates both Nrf2 accumulation and access to target genes.
Figures

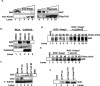

References
-
- Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a cap'n'collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 274:26071-26078. - PubMed
-
- Buetler, T. M., E. P. Gallagher, C. Wang, D. L. Stahl, J. D. Hayes, and D. L. Eaton. 1995. Induction of phase I and phase II drug-metabolizing enzyme mRNA, protein, and activity by BHA, ethoxyquin, and oltipraz. Toxicol. Appl. Pharmacol. 135:45-57. - PubMed
-
- Chan, J. Y., and M. Kwong. 2000. Impaired expression of glutathione synthetic enzyme genes in mice with targeted deletion of the Nrf2 basic-leucine zipper protein. Biochim. Biophys. Acta 1517:19-26. - PubMed
-
- Cullinan, S. B., and J. A. Diehl. 2004. PERK-dependent activation of Nrf2 contributes to redox homeostasis and cell survival following ER stress. J. Biol. Chem. 279:20076-20087. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
