Synapsis of recombination signal sequences located in cis and DNA underwinding in V(D)J recombination
- PMID: 15367690
- PMCID: PMC516766
- DOI: 10.1128/MCB.24.19.8727-8744.2004
Synapsis of recombination signal sequences located in cis and DNA underwinding in V(D)J recombination
Abstract
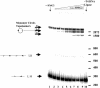
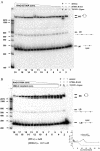
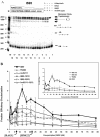
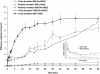
V(D)J recombination requires binding and synapsis of a complementary (12/23) pair of recombination signal sequences (RSSs) by the RAG1 and RAG2 proteins, aided by a high-mobility group protein, HMG1 or HMG2. Double-strand DNA cleavage within this synaptic, or paired, complex is thought to involve DNA distortion or melting near the site of cleavage. Although V(D)J recombination normally occurs between RSSs located on the same DNA molecule (in cis), all previous studies that directly assessed RSS synapsis were performed with the two DNA substrates in trans. To overcome this limitation, we have developed a facilitated circularization assay using DNA substrates of reduced length to assess synapsis of RSSs in cis. We show that a 12/23 pair of RSSs is the preferred substrate for synapsis of cis RSSs and that the efficiency of pairing is dependent upon RAG1-RAG2 stoichiometry. Synapsis in cis occurs rapidly and is kinetically favored over synapsis of RSSs located in trans. This experimental system also allowed the generation of underwound DNA substrates containing pairs of RSSs in cis. Importantly, we found that the RAG proteins cleave such substrates substantially more efficiently than relaxed substrates and that underwinding may enhance RSS synapsis as well as RAG1/2-mediated catalysis. The energy stored in such underwound substrates may be used in the generation of DNA distortion and/or protein conformational changes needed for synapsis and cleavage. We propose that this unwinding is uniquely sensed during synapsis of an appropriate 12/23 pair of RSSs.
Figures






Similar articles
-
HMGB1/2 can target DNA for illegitimate cleavage by the RAG1/2 complex.BMC Mol Biol. 2009 Mar 24;10:24. doi: 10.1186/1471-2199-10-24. BMC Mol Biol. 2009. PMID: 19317908 Free PMC article.
-
Full-length RAG1 promotes contact with coding and intersignal sequences in RAG protein complexes bound to recombination signals paired in cis.Nucleic Acids Res. 2009 Apr;37(7):2211-26. doi: 10.1093/nar/gkp047. Epub 2009 Feb 20. Nucleic Acids Res. 2009. PMID: 19233873 Free PMC article.
-
Fluorescence resonance energy transfer analysis of recombination signal sequence configuration in the RAG1/2 synaptic complex.Mol Cell Biol. 2007 Jul;27(13):4745-58. doi: 10.1128/MCB.00177-07. Epub 2007 Apr 30. Mol Cell Biol. 2007. PMID: 17470556 Free PMC article.
-
Early steps of V(D)J rearrangement: insights from biochemical studies of RAG-RSS complexes.Adv Exp Med Biol. 2009;650:1-15. doi: 10.1007/978-1-4419-0296-2_1. Adv Exp Med Biol. 2009. PMID: 19731797 Review.
-
V(D)J recombination: RAG proteins, repair factors, and regulation.Annu Rev Biochem. 2002;71:101-32. doi: 10.1146/annurev.biochem.71.090501.150203. Epub 2001 Nov 9. Annu Rev Biochem. 2002. PMID: 12045092 Review.
Cited by
-
Determinants of HMGB proteins required to promote RAG1/2-recombination signal sequence complex assembly and catalysis during V(D)J recombination.Mol Cell Biol. 2005 Jun;25(11):4413-25. doi: 10.1128/MCB.25.11.4413-4425.2005. Mol Cell Biol. 2005. PMID: 15899848 Free PMC article.
-
DNA bending in the synaptic complex in V(D)J recombination: turning an ancestral transpososome upside down.Discoveries (Craiova). 2014 Mar 29;2(1):e13. doi: 10.15190/d.2014.5. Discoveries (Craiova). 2014. PMID: 32309545 Free PMC article. Review.
-
Plasma HMGB-1 Levels in Subjects with Obesity and Type 2 Diabetes: A Cross-Sectional Study in China.PLoS One. 2015 Aug 28;10(8):e0136564. doi: 10.1371/journal.pone.0136564. eCollection 2015. PLoS One. 2015. PMID: 26317615 Free PMC article.
-
Bench-to-bedside review: High-mobility group box 1 and critical illness.Crit Care. 2007;11(5):229. doi: 10.1186/cc6088. Crit Care. 2007. PMID: 17903310 Free PMC article.
-
HMGB1, an alarmin promoting HIV dissemination and latency in dendritic cells.Cell Death Differ. 2012 Jan;19(1):96-106. doi: 10.1038/cdd.2011.134. Epub 2011 Oct 28. Cell Death Differ. 2012. PMID: 22033335 Free PMC article. Review.
References
-
- Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93:897-908. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
