Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis
- PMID: 15367692
- PMCID: PMC516737
- DOI: 10.1128/MCB.24.19.8753-8764.2004
Activity of the Yap1 transcription factor in Saccharomyces cerevisiae is modulated by methylglyoxal, a metabolite derived from glycolysis
Abstract
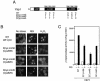
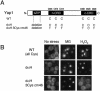
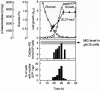
Methylglyoxal (MG) is synthesized during glycolysis, although it inhibits cell growth in all types of organisms. Hence, it has long been asked why such a toxic metabolite is synthesized in vivo. Glyoxalase I is a major enzyme detoxifying MG. Here we show that the Yap1 transcription factor, which is critical for the oxidative-stress response in Saccharomyces cerevisiae, is constitutively concentrated in the nucleus and activates the expression of its target genes in a glyoxalase I-deficient mutant. Yap1 contains six cysteine residues in two cysteine-rich domains (CRDs), i.e., three cysteine residues clustering near the N terminus (n-CRD) and the remaining three cysteine residues near the C terminus (c-CRD). We reveal that any of the three cysteine residues in the c-CRD is sufficient for MG to allow Yap1 to translocate into the nucleus and to activate the expression of its target gene. A Yap1 mutant possessing only one cysteine residue in the c-CRD but no cysteine in the n-CRD and deletion of the basic leucine zipper domain can concentrate in the nucleus with MG treatment. However, substitution of all the cysteine residues in Yap1 abolishes the ability of this transcription factor to concentrate in the nucleus following MG treatment. The redox status of Yap1 is substantially unchanged, and protein(s) interaction with Yap1 through disulfide bond is hardly detected in cells treated with MG. Collectively, neither intermolecular nor intramolecular disulfide bond formation seems to be involved in Yap1 activation by MG. Moreover, we show that nucleocytoplasmic localization of Yap1 closely correlates with growth phase and intracellular MG level. We propose a novel regulatory pathway underlying Yap1 activation by a natural metabolite in the cell.
Figures






References
-
- Abdulnur, S. F. 1976. The interactions of glyoxals with proteins and DNA in relation to cancer. Int. J. Quant. Chem. Quant. Biol. Symp. 3:59-64.
-
- Azevedo, D., F. Tacnet, A. Delaunay, C. Rodrigues-Pousada, and M. B. Toledano. 2003. Two redox centers within Yap1 for H2O2 and thiol-reactive chemicals signaling. Free Radic. Biol. Med. 35:889-900. - PubMed
-
- Bito, A., M. Haider, I. Hadler, and M. Breitenbach. 1997. Identification and phenotypic analysis of two glyoxalase II encoding genes from Saccharomyces cerevisiae, GLO2 and GLO4, and intracellular localization of the corresponding proteins. J. Biol. Chem. 272:21509-21519. - PubMed
-
- Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. - PubMed
-
- Carmel-Harel, O., R. Stearman, A. P. Gasch, D. Botstein, P. O. Brown, and G. Storz. 2001. Role of thioredoxin reductase in the Yap1-dependent response to oxidative stress in Saccharomyces cerevisiae. Mol. Microbiol. 39:595-605. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
