DNA gyrase is involved in chloroplast nucleoid partitioning
- PMID: 15367714
- PMCID: PMC520963
- DOI: 10.1105/tpc.104.024281
DNA gyrase is involved in chloroplast nucleoid partitioning
Abstract
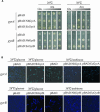
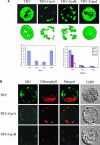
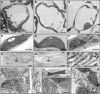
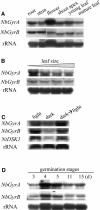
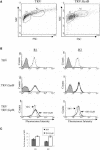
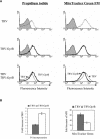
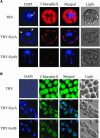
DNA gyrase, which catalyzes topological transformation of DNA, plays an essential role in replication and transcription in prokaryotes. Virus-induced gene silencing of NbGyrA or NbGyrB, which putatively encode DNA gyrase subunits A and B, respectively, resulted in leaf yellowing phenotypes in Nicotiana benthamiana. NbGyrA and NbGyrB complemented the gyrA and gyrB temperature-sensitive mutations of Escherichia coli, respectively, which indicates that the plant and bacterial subunits are functionally similar. NbGyrA and NbGyrB were targeted to both chloroplasts and mitochondria, and depletion of these subunits affected both organelles by reducing chloroplast numbers and inducing morphological and physiological abnormalities in both organelles. Flow cytometry analysis revealed that the average DNA content in the affected chloroplasts and mitochondria was significantly higher than in the control organelles. Furthermore, 4',6-diamidino-2-phenylindole staining revealed that the abnormal chloroplasts contained one or a few large nucleoids instead of multiple small nucleoids dispersed throughout the stroma. Pulse-field gel electrophoresis analyses of chloroplasts demonstrated that the sizes and/or structure of the DNA molecules in the abnormal chloroplast nucleoids are highly aberrant. Based on these results, we propose that DNA gyrase plays a critical role in chloroplast nucleoid partitioning by regulating DNA topology.
Figures




References
-
- Champoux, J.J. (2001). DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413. - PubMed
-
- Cho, H.S., Yoon, G.M., Lee, S.S., Kim, Y.A., Hwang, I., Choi, D., and Pai, H.S. (2001). A novel dual-specificity protein kinase targeted to the chloroplast in tobacco. FEBS Lett. 497, 124–130. - PubMed
-
- Corbett, K.D., and Berger, J.D. (2003). Emerging roles for plant topoisomerase VI. Chem. Biol. 10, 107–111. - PubMed
-
- Douce, R., Bourguignon, J., Brouquisse, R., and Neuburger, M. (1987). Isolation of plant mitochondria. Methods Enzymol. 148, 403–415.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

