Hormonal regulation of plant growth and development
- PMID: 15367944
- PMCID: PMC516799
- DOI: 10.1371/journal.pbio.0020311
Hormonal regulation of plant growth and development
Abstract
Besides environmental factors, plant growth depends upon endogenous signals. Bill Gray examines what these hormonal signals are and how they act to regulate many aspects of growth and development.
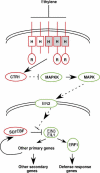
Figures


References
-
- Chang C. Ethylene signaling: The MAPK module has finally landed. Trends Plant Sci. 2003;8:365–368. - PubMed
-
- Deshaies RJ. SCF and Cullin/Ring H2-based ubiquitin ligases. Annu Rev Cell Dev Biol. 1999;15:435–467. - PubMed
-
- Dharmasiri N, Estelle M. Auxin signaling and regulated protein degradation. Trends Plant Sci. 2004;9:302–308. - PubMed
-
- Gomi K, Matsuoka M. Gibberellin signalling pathway. Curr Opin Plant Biol. 2003;6:489–493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

