Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae
- PMID: 15371350
- PMCID: PMC1448728
- DOI: 10.1534/genetics.104.033266
Haploidy, diploidy and evolution of antifungal drug resistance in Saccharomyces cerevisiae
Abstract
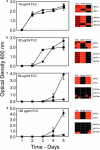
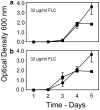
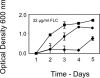
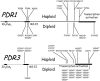
We tested the hypothesis that the time course of the evolution of antifungal drug resistance depends on the ploidy of the fungus. The experiments were designed to measure the initial response to the selection imposed by the antifungal drug fluconazole up to and including the fixation of the first resistance mutation in populations of Saccharomyces cerevisiae. Under conditions of low drug concentration, mutations in the genes PDR1 and PDR3, which regulate the ABC transporters implicated in resistance to fluconazole, are favored. In this environment, diploid populations of defined size consistently became fixed for a resistance mutation sooner than haploid populations. Experiments manipulating population sizes showed that this advantage of diploids was due to increased mutation availability relative to that of haploids; in effect, diploids have twice the number of mutational targets as haploids and hence have a reduced waiting time for mutations to occur. Under conditions of high drug concentration, recessive mutations in ERG3, which result in resistance through altered sterol synthesis, are favored. In this environment, haploids consistently achieved resistance much sooner than diploids. When 29 haploid and 29 diploid populations were evolved for 100 generations in low drug concentration, the mutations fixed in diploid populations were all dominant, while the mutations fixed in haploid populations were either recessive (16 populations) or dominant (13 populations). Further, the spectrum of the 53 nonsynonymous mutations identified at the sequence level was different between haploids and diploids. These results fit existing theory on the relative abilities of haploids and diploids to adapt and suggest that the ploidy of the fungal pathogen has a strong impact on the evolution of fluconazole resistance.
Figures
Similar articles
-
Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae.Genetics. 2003 Apr;163(4):1287-98. doi: 10.1093/genetics/163.4.1287. Genetics. 2003. PMID: 12702675 Free PMC article.
-
Antagonism between two mechanisms of antifungal drug resistance.Eukaryot Cell. 2006 Aug;5(8):1243-51. doi: 10.1128/EC.00048-06. Eukaryot Cell. 2006. PMID: 16896209 Free PMC article.
-
The limit to evolutionary rescue depends on ploidy in yeast exposed to nystatin.Can J Microbiol. 2024 Sep 1;70(9):394-404. doi: 10.1139/cjm-2023-0235. Epub 2024 Jun 14. Can J Microbiol. 2024. PMID: 38875715
-
Experimental studies on ploidy evolution in yeast.FEMS Microbiol Lett. 2004 Apr 15;233(2):187-92. doi: 10.1111/j.1574-6968.2004.tb09481.x. FEMS Microbiol Lett. 2004. PMID: 15108721 Review.
-
Evolution of antifungal-drug resistance: mechanisms and pathogen fitness.Nat Rev Microbiol. 2005 Jul;3(7):547-56. doi: 10.1038/nrmicro1179. Nat Rev Microbiol. 2005. PMID: 15953931 Review.
Cited by
-
Genomic convergence toward diploidy in Saccharomyces cerevisiae.PLoS Genet. 2006 Sep 22;2(9):e145. doi: 10.1371/journal.pgen.0020145. PLoS Genet. 2006. PMID: 17002497 Free PMC article.
-
Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance.PLoS Genet. 2009 Oct;5(10):e1000705. doi: 10.1371/journal.pgen.1000705. Epub 2009 Oct 30. PLoS Genet. 2009. PMID: 19876375 Free PMC article.
-
Antifungal Drug Concentration Impacts the Spectrum of Adaptive Mutations in Candida albicans.Mol Biol Evol. 2023 Jan 4;40(1):msad009. doi: 10.1093/molbev/msad009. Mol Biol Evol. 2023. PMID: 36649220 Free PMC article.
-
Sensory input attenuation allows predictive sexual response in yeast.Nat Commun. 2016 Aug 25;7:12590. doi: 10.1038/ncomms12590. Nat Commun. 2016. PMID: 27557894 Free PMC article.
-
Genomic Diversity across Candida auris Clinical Isolates Shapes Rapid Development of Antifungal Resistance In Vitro and In Vivo.mBio. 2022 Aug 30;13(4):e0084222. doi: 10.1128/mbio.00842-22. Epub 2022 Jul 5. mBio. 2022. PMID: 35862787 Free PMC article.
References
-
- Berman, J., and P. E. Sudbery, 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3: 918–930. - PubMed
-
- Kolaczkowska, A., M. Kolaczkowski, A. Delahodde and A. Goffeau, 2002. Functional dissection of Pdr1p, a regulator of multidrug resistance in Saccharomyces cerevisiae. Mol. Genet. Genomics 267: 96–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

