The complex flagellar torque generator of Pseudomonas aeruginosa
- PMID: 15375113
- PMCID: PMC516612
- DOI: 10.1128/JB.186.19.6341-6350.2004
The complex flagellar torque generator of Pseudomonas aeruginosa
Abstract
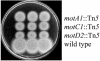
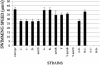
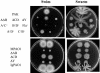
Flagella act as semirigid helical propellers that are powered by reversible rotary motors. Two membrane proteins, MotA and MotB, function as a complex that acts as the stator and generates the torque that drives rotation. The genome sequence of Pseudomonas aeruginosa PAO1 contains dual sets of motA and motB genes, PA1460-PA1461 (motAB) and PA4954-PA4953 (motCD), as well as another gene, motY (PA3526), which is known to be required for motor function in some bacteria. Here, we show that these five genes contribute to motility. Loss of function of either motAB-like locus was dispensable for translocation in aqueous environments. However, swimming could be entirely eliminated by introduction of combinations of mutations in the two motAB-encoding regions. Mutation of both genes encoding the MotA homologs or MotB homologs was sufficient to abolish motility. Mutants carrying double mutations in nonequivalent genes (i.e., motA motD or motB motC) retained motility, indicating that noncognate components can function together. motY appears to be required for motAB function. The combination of motY and motCD mutations rendered the cells nonmotile. Loss of function of motAB, motY, or motAB motY produced similar phenotypes; although the swimming speed was only reduced to approximately 85% of the wild-type speed, translocation in semisolid motility agar and swarming on the surface of solidified agar were severely impeded. Thus, the flagellar motor of P. aeruginosa represents a more complex configuration than the configuration that has been studied in other bacteria, and it enables efficient movement under different circumstances.
Figures

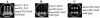



References
-
- Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. - PubMed
-
- Armitage, J. P. 1999. Bacterial tactic responses. Adv. Microb. Physiol. 41:229-289. - PubMed
-
- Berg, H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19-54. - PubMed
-
- Blair, D. F. 2003. Flagellar movement driven by proton translocation. FEBS Lett. 545:86-95. - PubMed
-
- Blair, D. F., and H. C. Berg. 1988. Restoration of torque in defective flagellar motors. Science 242:1678-1681. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

