Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system
- PMID: 15375120
- PMCID: PMC516608
- DOI: 10.1128/JB.186.19.6400-6408.2004
Antisense RNA regulation by stable complex formation in the Enterococcus faecalis plasmid pAD1 par addiction system
Abstract
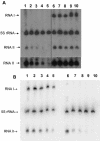
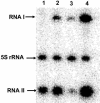
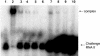
The par stability determinant, encoded by the Enterococcus faecalis plasmid pAD1, is the only antisense RNA regulated postsegregational killing system identified in gram-positive bacteria. Because of the unique organization of the par locus, the par antisense RNA, RNA II, binds to its target, RNA I, at relatively small, interspersed regions of complementarity. The results of this study suggest that, rather than targeting the antisense bound message for rapid degradation, as occurs in most other antisense RNA regulated systems, RNA I and RNA II form a relatively stable, presumably translationally inactive complex. The stability of the RNA I-RNA II complex would allow RNA I to persist in an untranslated state unless or until the encoding plasmid was lost. After plasmid loss, RNA II would be removed from the complex, allowing translational activation of RNA I. The mechanism of RNA I activation in vivo is unknown, but in vitro dissociation experiments suggest that active removal of RNA II, for example by a cellular RNase, may be required.
Figures
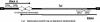

Similar articles
-
The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs.RNA Biol. 2012 Dec;9(12):1498-503. doi: 10.4161/rna.22311. Epub 2012 Oct 11. RNA Biol. 2012. PMID: 23059908 Free PMC article. Review.
-
Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI.Mol Microbiol. 2001 Oct;42(2):527-37. doi: 10.1046/j.1365-2958.2001.02663.x. Mol Microbiol. 2001. PMID: 11703673
-
An intramolecular upstream helix ensures the stability of a toxin-encoding RNA in Enterococcus faecalis.J Bacteriol. 2009 Mar;191(5):1528-36. doi: 10.1128/JB.01316-08. Epub 2008 Dec 19. J Bacteriol. 2009. PMID: 19103923 Free PMC article.
-
Antisense RNA regulation of the pAD1 par post-segregational killing system requires interaction at the 5' and 3' ends of the RNAs.Mol Microbiol. 2000 Aug;37(3):661-70. doi: 10.1046/j.1365-2958.2000.02034.x. Mol Microbiol. 2000. PMID: 10931359
-
The Type I toxin-antitoxin par locus from Enterococcus faecalis plasmid pAD1: RNA regulation by both cis- and trans-acting elements.Plasmid. 2015 Mar;78:65-70. doi: 10.1016/j.plasmid.2014.10.001. Epub 2014 Oct 13. Plasmid. 2015. PMID: 25312777 Free PMC article. Review.
Cited by
-
Modeling sRNA-Regulated Plasmid Maintenance.PLoS One. 2017 Jan 13;12(1):e0169703. doi: 10.1371/journal.pone.0169703. eCollection 2017. PLoS One. 2017. PMID: 28085919 Free PMC article.
-
Type I toxin-antitoxin systems in bacteria: from regulation to biological functions.EcoSal Plus. 2024 Dec 12;12(1):eesp00252022. doi: 10.1128/ecosalplus.esp-0025-2022. Epub 2024 May 20. EcoSal Plus. 2024. PMID: 38767346 Free PMC article. Review.
-
The par toxin-antitoxin system from Enterococcus faecalis plasmid pAD1 and its chromosomal homologs.RNA Biol. 2012 Dec;9(12):1498-503. doi: 10.4161/rna.22311. Epub 2012 Oct 11. RNA Biol. 2012. PMID: 23059908 Free PMC article. Review.
-
Plasmid addiction systems: perspectives and applications in biotechnology.Microb Biotechnol. 2010 Nov;3(6):634-57. doi: 10.1111/j.1751-7915.2010.00170.x. Microb Biotechnol. 2010. PMID: 21255361 Free PMC article. Review.
-
sRNA antitoxins: more than one way to repress a toxin.Toxins (Basel). 2014 Aug 4;6(8):2310-35. doi: 10.3390/toxins6082310. Toxins (Basel). 2014. PMID: 25093388 Free PMC article. Review.
References
-
- Andersen, J., S. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. - PubMed
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Boston, Mass.
-
- Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

