Insecticidal pilin subunit from the insect pathogen Xenorhabdus nematophila
- PMID: 15375127
- PMCID: PMC516617
- DOI: 10.1128/JB.186.19.6465-6476.2004
Insecticidal pilin subunit from the insect pathogen Xenorhabdus nematophila
Abstract
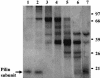
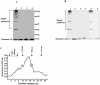
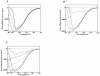
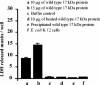
Xenorhabdus nematophila is an insect pathogen and produces protein toxins which kill the larval host. Previously, we characterized an orally toxic, large, outer membrane-associated protein complex from the culture medium of X. nematophila. Here, we describe the cloning, expression, and characterization of a 17-kDa pilin subunit of X. nematophila isolated from that protein complex. The gene was amplified by PCR, cloned, and expressed in Escherichia coli. The recombinant protein was refolded in vitro in the absence of its cognate chaperone by using a urea gradient. The protein oligomerized during in vitro refolding, forming multimers. Point mutations in the conserved N-terminal residues of the pilin protein greatly destabilized its oligomeric organization, demonstrating the importance of the N terminus in refolding and oligomerization of the pilin subunit by donor strand complementation. The recombinant protein was cytotoxic to cultured Helicoverpa armigera larval hemocytes, causing agglutination and subsequent release of the cytoplasmic enzyme lactate dehydrogenase. The agglutination of larval cells by the 17-kDa protein was inhibited by several sugar derivatives. The biological activity of the purified recombinant protein indicated that it has a conformation similar to that of the native protein. The 17-kDa pilin subunit was found to be orally toxic to fourth- or fifth-instar larvae of an important crop pest, H. armigera, causing extensive damage to the midgut epithelial membrane. To our knowledge, this is first report describing an insecticidal pilin subunit of a bacterium.
Figures





Similar articles
-
The cytotoxic fimbrial structural subunit of Xenorhabdus nematophila is a pore-forming toxin.J Bacteriol. 2006 Nov;188(22):7957-62. doi: 10.1128/JB.00787-06. Epub 2006 Sep 1. J Bacteriol. 2006. PMID: 16950919 Free PMC article.
-
Characterization of a cytotoxic pilin subunit of Xenorhabdus nematophila.Biochem Biophys Res Commun. 2004 Feb 20;314(4):943-9. doi: 10.1016/j.bbrc.2003.12.187. Biochem Biophys Res Commun. 2004. PMID: 14751223
-
A novel pilin subunit from Xenorhabdus nematophila, an insect pathogen, confers pest resistance in tobacco and tomato.Plant Cell Rep. 2015 Nov;34(11):1863-72. doi: 10.1007/s00299-015-1833-6. Epub 2015 Jul 12. Plant Cell Rep. 2015. PMID: 26164296
-
Novel insecticidal chitinase from the insect pathogen Xenorhabdus nematophila.Int J Biol Macromol. 2020 Sep 15;159:394-401. doi: 10.1016/j.ijbiomac.2020.05.078. Epub 2020 May 15. Int J Biol Macromol. 2020. PMID: 32422264
-
[Purification and characterization of the haemocoel insecticide Tp40 from Xenorhabdus nematophila].Wei Sheng Wu Xue Bao. 2008 May;48(5):677-83. Wei Sheng Wu Xue Bao. 2008. PMID: 18652303 Chinese.
Cited by
-
Recombinant entomopathogenic agents: a review of biotechnological approaches to pest insect control.World J Microbiol Biotechnol. 2017 Dec 18;34(1):14. doi: 10.1007/s11274-017-2397-0. World J Microbiol Biotechnol. 2017. PMID: 29255969 Review.
-
Bacteroides: the good, the bad, and the nitty-gritty.Clin Microbiol Rev. 2007 Oct;20(4):593-621. doi: 10.1128/CMR.00008-07. Clin Microbiol Rev. 2007. PMID: 17934076 Free PMC article. Review.
-
The cytotoxic fimbrial structural subunit of Xenorhabdus nematophila is a pore-forming toxin.J Bacteriol. 2006 Nov;188(22):7957-62. doi: 10.1128/JB.00787-06. Epub 2006 Sep 1. J Bacteriol. 2006. PMID: 16950919 Free PMC article.
-
Functional Characterization of Outer Membrane Proteins (OMPs) in Xenorhabdus nematophila and Photorhabdus luminescens through Insect Immune Defense Reactions.Insects. 2019 Oct 17;10(10):352. doi: 10.3390/insects10100352. Insects. 2019. PMID: 31627300 Free PMC article.
-
An insecticidal GroEL protein with chitin binding activity from Xenorhabdus nematophila.J Biol Chem. 2008 Oct 17;283(42):28287-96. doi: 10.1074/jbc.M804416200. Epub 2008 Jul 30. J Biol Chem. 2008. PMID: 18667427 Free PMC article.
References
-
- Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp. bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditae and Steinernematidae. J. Gen. Microbiol. 128:3061-3065. - PubMed
-
- Akhurst, R. J., and G. B. Dunphy. 1993. Symbiotically associated entomopathogenic bacteria, nematodes and their insect hosts, p. 1-23. In N. Beckage, S. Thompson, and B. Federici (ed.), Parasites and pathogens of insects, vol. 2. Academic Press, Inc., New York, N.Y.
-
- Boemare, N. E., and R. J. Akhrust. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp. ( Enteriobacteriaceae). J. Gen. Microbiol. 134:751-761.
-
- Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. Ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

