Bifidobacterium longum requires a fructokinase (Frk; ATP:D-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism
- PMID: 15375133
- PMCID: PMC516584
- DOI: 10.1128/JB.186.19.6515-6525.2004
Bifidobacterium longum requires a fructokinase (Frk; ATP:D-fructose 6-phosphotransferase, EC 2.7.1.4) for fructose catabolism
Abstract
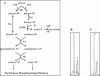
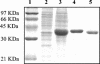
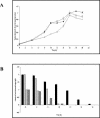
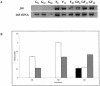
Although the ability of Bifidobacterium spp. to grow on fructose as a unique carbon source has been demonstrated, the enzyme(s) needed to incorporate fructose into a catabolic pathway has hitherto not been defined. This work demonstrates that intracellular fructose is metabolized via the fructose-6-P phosphoketolase pathway and suggests that a fructokinase (Frk; EC 2.7.1.4) is the enzyme that is necessary and sufficient for the assimilation of fructose into this catabolic route in Bifidobacterium longum. The B. longum A10C fructokinase-encoding gene (frk) was expressed in Escherichia coli from a pET28 vector with an attached N-terminal histidine tag. The expressed enzyme was purified by affinity chromatography on a Co(2+)-based column, and the pH and temperature optima were determined. A biochemical analysis revealed that Frk displays the same affinity for fructose and ATP (Km(fructose) = 0.739 +/- 0.18 mM and Km(ATP) = 0.756 +/- 0.08 mM), is highly specific for D-fructose, and is inhibited by an excess of ATP (>12 mM). It was also found that frk is inducible by fructose and is subject to glucose-mediated repression. Consequently, this work presents the first characterization at the molecular and biochemical level of a fructokinase from a gram-positive bacterium that is highly specific for D-fructose.
Figures



Similar articles
-
Cloning, sequencing, and expression of the Zymomonas mobilis fructokinase gene and structural comparison of the enzyme with other hexose kinases.J Bacteriol. 1992 Jun;174(11):3455-60. doi: 10.1128/jb.174.11.3455-3460.1992. J Bacteriol. 1992. PMID: 1317376 Free PMC article.
-
Identification and biochemical characterization of the fructokinase gene family in Arabidopsis thaliana.BMC Plant Biol. 2017 Apr 26;17(1):83. doi: 10.1186/s12870-017-1031-5. BMC Plant Biol. 2017. PMID: 28441933 Free PMC article.
-
Characterisation of glutamine fructose-6-phosphate amidotransferase (EC 2.6.1.16) and N-acetylglucosamine metabolism in Bifidobacterium.Arch Microbiol. 2008 Feb;189(2):157-67. doi: 10.1007/s00203-007-0307-9. Epub 2007 Oct 18. Arch Microbiol. 2008. PMID: 17943273
-
Plant Fructokinases: Evolutionary, Developmental, and Metabolic Aspects in Sink Tissues.Front Plant Sci. 2018 Mar 16;9:339. doi: 10.3389/fpls.2018.00339. eCollection 2018. Front Plant Sci. 2018. PMID: 29616058 Free PMC article. Review.
-
The anhydrofructose pathway and its possible role in stress response and signaling.Biochim Biophys Acta. 2006 Sep;1760(9):1314-22. doi: 10.1016/j.bbagen.2006.05.007. Epub 2006 Jun 2. Biochim Biophys Acta. 2006. PMID: 16822618 Review.
Cited by
-
Lactose-over-glucose preference in Bifidobacterium longum NCC2705: glcP, encoding a glucose transporter, is subject to lactose repression.J Bacteriol. 2006 Feb;188(4):1260-5. doi: 10.1128/JB.188.4.1260-1265.2006. J Bacteriol. 2006. PMID: 16452407 Free PMC article.
-
Mannheimia succiniciproducens phosphotransferase system for sucrose utilization.Appl Environ Microbiol. 2010 Mar;76(5):1699-703. doi: 10.1128/AEM.02468-09. Epub 2010 Jan 15. Appl Environ Microbiol. 2010. PMID: 20081002 Free PMC article.
-
Proteomic Signatures of Microbial Adaptation to the Highest Ultraviolet-Irradiation on Earth: Lessons From a Soil Actinobacterium.Front Microbiol. 2022 Mar 15;13:791714. doi: 10.3389/fmicb.2022.791714. eCollection 2022. Front Microbiol. 2022. PMID: 35369494 Free PMC article.
-
Deciphering the role of recurrent FAD-dependent enzymes in bacterial phosphonate catabolism.iScience. 2023 Oct 4;26(11):108108. doi: 10.1016/j.isci.2023.108108. eCollection 2023 Nov 17. iScience. 2023. PMID: 37876809 Free PMC article.
-
The Microbial Degradation of Natural and Anthropogenic Phosphonates.Molecules. 2023 Sep 29;28(19):6863. doi: 10.3390/molecules28196863. Molecules. 2023. PMID: 37836707 Free PMC article. Review.
References
-
- Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. - PubMed
-
- Aulkemeyer, P., R. Ebner, G. Heilenmann, K. Jahreis, K. Schmid, S. Wrieden, and J. W. Lengeler. 1991. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol. Microbiol. 5:2913-2922. - PubMed
-
- Berghauser, J. 1975. A reactive arginine in adenylate kinase. Biochim. Biophys. Acta 397:370-376. - PubMed
-
- Bockmann, J., H. Heuel, and J. W. Lengeler. 1992. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. 235:22-32. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

