Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence
- PMID: 15375142
- PMCID: PMC516600
- DOI: 10.1128/JB.186.19.6605-6616.2004
Transcription regulation by the Mycobacterium tuberculosis alternative sigma factor SigD and its role in virulence
Abstract
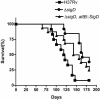
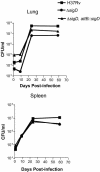
Mycobacterium tuberculosis, an obligate mammalian pathogen, adapts to its host during the course of infection via the regulation of gene expression. Of the regulators of transcription that play a role in this response, several alternative sigma factors of M. tuberculosis have been shown to control gene expression in response to stresses, and some of these are required for virulence or persistence in vivo. For this study, we examined the role of the alternative sigma factor SigD in M. tuberculosis gene expression and virulence. Using microarray analysis, we identified several genes whose expression was altered in a strain with a sigD deletion. A small number of these genes, including sigD itself, the gene encoding the autocrine growth factor RpfC, and a gene of unknown function, Rv1815, appear to be directly regulated by this sigma factor. By identifying the in vivo promoters of these genes, we have determined a consensus promoter sequence that is putatively recognized by SigD. The expression of several genes encoding PE-PGRS proteins, part of a large family of related genes of unknown function, was significantly increased in the sigD mutant. We found that the expression of sigD is stable throughout log phase and stationary phase but that it declines rapidly with oxygen depletion. In a mouse infection model, the sigD mutant strain was attenuated, with differences in survival and the inflammatory response in the lung between mice infected with the mutant and those infected with the wild type.
Figures

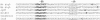

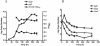

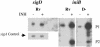

References
-
- Banu, S., N. Honore, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44:9-19. - PubMed
-
- Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

