Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing
- PMID: 15375210
- PMCID: PMC521097
- DOI: 10.1073/pnas.0402745101
Genetic exchange and plasmid transfers in Borrelia burgdorferi sensu stricto revealed by three-way genome comparisons and multilocus sequence typing
Abstract
Comparative genomics of closely related bacterial isolates is a powerful method for uncovering virulence and other important genome elements. We determined draft sequences (8-fold coverage) of the genomes of strains JD1 and N40 of Borrelia burgdorferi sensu stricto, the causative agent of Lyme disease, and we compared the predicted genes from the two genomes with those from the previously sequenced B31 genome. The three genomes are closely related and are evolutionarily approximately equidistant ( approximately 0.5% pairwise nucleotide differences on the main chromosome). We used a Poisson model of nucleotide substitution to screen for genes with elevated levels of nucleotide polymorphisms. The three-way genome comparison allowed distinction between polymorphisms introduced by mutations and those introduced by recombination using the method of phylogenetic partitioning. Tests for recombination suggested that patches of high-density nucleotide polymorphisms on the chromosome and plasmids arise by DNA exchange. The role of recombination as the main mechanism driving B. burgdorferi diversification was confirmed by multilocus sequence typing of 18 clinical isolates at 18 polymorphic loci. A strong linkage between the multilocus sequence genotypes and the major alleles of outer-surface protein C (ospC) suggested that balancing selection at ospC is a dominant force maintaining B. burgdorferi diversity in local populations. We conclude that B. burgdorferi undergoes genome-wide genetic exchange, including plasmid transfers, and previous reports of its clonality are artifacts from the use of geographically and ecological isolated samples. Frequent recombination implies a potential for rapid adaptive evolution and a possible polygenic basis of B. burgdorferi pathogenicity.
Figures
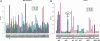

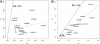
References
-
- Read, T. D., Salzberg, S. L., Pop, M., Shumway, M., Umayam, L., Jiang, L., Holtzapple, E., Busch, J. D., Smith, K. L., Schupp, J. M., et al. (2002) Science 296, 2028–2033. - PubMed
-
- Perna, N. T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J. D., Rose, D. J., Mayhew, G. F., Evans, P. S., Gregor, J., Kirkpatrick, H. A., et al. (2001) Nature 409, 529–533. - PubMed
-
- Read, T. D., Peterson, S. N., Tourasse, N., Baillie, L. W., Paulsen, I. T., Nelson, K. E., Tettelin, H., Fouts, D. E., Eisen, J. A., Gill, S. R., et al. (2003) Nature 423, 81–86. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

