Identification of mushroom body miniature, a zinc-finger protein implicated in brain development of Drosophila
- PMID: 15375215
- PMCID: PMC521146
- DOI: 10.1073/pnas.0405887101
Identification of mushroom body miniature, a zinc-finger protein implicated in brain development of Drosophila
Abstract
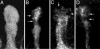
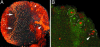
The mushroom bodies are bilaterally arranged structures in the protocerebrum of Drosophila and most other insect species. Mutants with altered mushroom body structure have been instrumental not only in establishing their role in distinct behavioral functions but also in identifying the molecular pathways that control mushroom body development. The mushroom body miniature(1) (mbm(1)) mutation results in grossly reduced mushroom bodies and odor learning deficits in females. With a survey of genomic rescue constructs, we have pinpointed mbm(1) to a single transcription unit and identified a single nucleotide exchange in the 5' untranslated region of the corresponding transcript resulting in a reduced expression of the protein. The most obvious feature of the Mbm protein is a pair of C(2)HC zinc fingers, implicating a function of the protein in binding nucleic acids. Immunohistochemical analysis shows that expression of the Mbm protein is not restricted to the mushroom bodies. BrdUrd labeling experiments indicate a function of Mbm in neuronal precursor cell proliferation.
Figures



Similar articles
-
Gradients of the Drosophila Chinmo BTB-zinc finger protein govern neuronal temporal identity.Cell. 2006 Oct 20;127(2):409-22. doi: 10.1016/j.cell.2006.08.045. Cell. 2006. PMID: 17055440
-
Structural brain mutants: mushroom body defect (mud): a case study.J Neurogenet. 2009;23(1-2):42-7. doi: 10.1080/01677060802471700. Epub 2008 Dec 23. J Neurogenet. 2009. PMID: 19107630 Review.
-
Organization of the honey bee mushroom body: representation of the calyx within the vertical and gamma lobes.J Comp Neurol. 2002 Aug 12;450(1):4-33. doi: 10.1002/cne.10285. J Comp Neurol. 2002. PMID: 12124764
-
Characterization of the dCaMKII-GAL4 driver line whose expression is controlled by the Drosophila Ca2+/calmodulin-dependent protein kinase II promoter.Cell Tissue Res. 2002 Nov;310(2):237-52. doi: 10.1007/s00441-002-0631-y. Epub 2002 Sep 28. Cell Tissue Res. 2002. PMID: 12397378
-
Regulation and function of the terminal gap gene huckebein in the Drosophila blastoderm.Int J Dev Biol. 1996 Feb;40(1):157-65. Int J Dev Biol. 1996. PMID: 8735925 Review.
Cited by
-
Drosophila mbm is a nucleolar myc and casein kinase 2 target required for ribosome biogenesis and cell growth of central brain neuroblasts.Mol Cell Biol. 2014 May;34(10):1878-91. doi: 10.1128/MCB.00658-13. Epub 2014 Mar 10. Mol Cell Biol. 2014. PMID: 24615015 Free PMC article.
-
Tip60 HAT Action Mediates Environmental Enrichment Induced Cognitive Restoration.PLoS One. 2016 Jul 25;11(7):e0159623. doi: 10.1371/journal.pone.0159623. eCollection 2016. PLoS One. 2016. PMID: 27454757 Free PMC article.
-
The unfulfilled gene is required for the development of mushroom body neuropil in Drosophila.Neural Dev. 2010 Feb 1;5:4. doi: 10.1186/1749-8104-5-4. Neural Dev. 2010. PMID: 20122139 Free PMC article.
-
Ten-a affects the fusion of central complex primordia in Drosophila.PLoS One. 2013;8(2):e57129. doi: 10.1371/journal.pone.0057129. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437330 Free PMC article.
-
The nsdC gene encoding a putative C2H2-type transcription factor is a key activator of sexual development in Aspergillus nidulans.Genetics. 2009 Jul;182(3):771-83. doi: 10.1534/genetics.109.101667. Epub 2009 May 4. Genetics. 2009. PMID: 19416940 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

