Reconstruction of ancestral protein sequences and its applications
- PMID: 15377393
- PMCID: PMC522809
- DOI: 10.1186/1471-2148-4-33
Reconstruction of ancestral protein sequences and its applications
Abstract
Background: Modern-day proteins were selected during long evolutionary history as descendants of ancient life forms. In silico reconstruction of such ancestral protein sequences facilitates our understanding of evolutionary processes, protein classification and biological function. Additionally, reconstructed ancestral protein sequences could serve to fill in sequence space thus aiding remote homology inference.
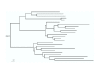
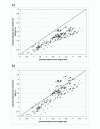
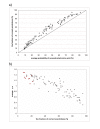
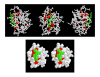
Results: We developed ANCESCON, a package for distance-based phylogenetic inference and reconstruction of ancestral protein sequences that takes into account the observed variation of evolutionary rates between positions that more precisely describes the evolution of protein families. To improve the accuracy of evolutionary distance estimation and ancestral sequence reconstruction, two approaches are proposed to estimate position-specific evolutionary rates. Comparisons show that at large evolutionary distances our method gives more accurate ancestral sequence reconstruction than PAML, PHYLIP and PAUP*. We apply the reconstructed ancestral sequences to homology inference and functional site prediction. We show that the usage of hypothetical ancestors together with the present day sequences improves profile-based sequence similarity searches; and that ancestral sequence reconstruction methods can be used to predict positions with functional specificity.
Conclusions: As a computational tool to reconstruct ancestral protein sequences from a given multiple sequence alignment, ANCESCON shows high accuracy in tests and helps detection of remote homologs and prediction of functional sites. ANCESCON is freely available for non-commercial use. Pre-compiled versions for several platforms can be downloaded from ftp://iole.swmed.edu/pub/ANCESCON/.
Figures






References
-
- Fitch WM. Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool. 1971;20:406–416.
-
- Hartigan JA. Minimum evolution fits to a given tree. Biometrics. 1973;29:53–65.
-
- Koshi JM, Goldstein RA. Probabilistic reconstruction of ancestral protein sequences. J Mol Evol. 1996;42:313–320. - PubMed
-
- Pupko T, Pe'er I, Shamir R, Graur D. A fast algorithm for joint reconstruction of ancestral amino acid sequences. Mol Biol Evol. 2000;17:890–896. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

