Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism
- PMID: 15377757
- PMCID: PMC520969
- DOI: 10.1105/tpc.104.023705
Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism
Abstract
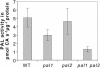
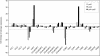
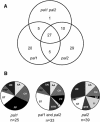
The first enzyme of the phenylpropanoid pathway, Phe ammonia-lyase (PAL), is encoded by four genes in Arabidopsis thaliana. Whereas PAL function is well established in various plants, an insight into the functional significance of individual gene family members is lacking. We show that in the absence of clear phenotypic alterations in the Arabidopsis pal1 and pal2 single mutants and with limited phenotypic alterations in the pal1 pal2 double mutant, significant modifications occur in the transcriptome and metabolome of the pal mutants. The disruption of PAL led to transcriptomic adaptation of components of the phenylpropanoid biosynthesis, carbohydrate metabolism, and amino acid metabolism, revealing complex interactions at the level of gene expression between these pathways. Corresponding biochemical changes included a decrease in the three major flavonol glycosides, glycosylated vanillic acid, scopolin, and two novel feruloyl malates coupled to coniferyl alcohol. Moreover, Phe overaccumulated in the double mutant, and the levels of many other amino acids were significantly imbalanced. The lignin content was significantly reduced, and the syringyl/guaiacyl ratio of lignin monomers had increased. Together, from the molecular phenotype, common and specific functions of PAL1 and PAL2 are delineated, and PAL1 is qualified as being more important for the generation of phenylpropanoids.
Figures




References
-
- Allwood, E.G., Davies, D.R., Gerrish, C., Ellis, B.E., and Bolwell, G.P. (1999). Phosphorylation of phenylalanine ammonia-lyase: Evidence for a novel protein kinase and identification of the phosphorylated residue. FEBS Lett. 457, 47–52. - PubMed
-
- Anterola, A.M., Jeon, J.-H., Davin, L.B., and Lewis, N.G. (2002). Transcriptional control of monolignol biosynthesis in Pinus taeda: Factors affecting monolignol ratios and carbon allocation in phenylpropanoid metabolism. J. Biol. Chem. 277, 18272–18280. - PubMed
-
- Bate, N.J., Orr, J., Ni, W., Meromi, A., Nadler-Hassar, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91, 7608–7612. - PMC - PubMed
-
- Baumert, A., Mock, H.-P., Schmidt, J., Herbers, K., Sonnewald, U., and Strack, D. (2001). Patterns of phenylpropanoids in non-inoculated and potato virus Y-inoculated leaves of transgenic tobacco plants expressing yeast-derived invertase. Phytochemistry 56, 535–541. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

