An experimentally determined protein folding energy landscape
- PMID: 15377792
- PMCID: PMC521126
- DOI: 10.1073/pnas.0403386101
An experimentally determined protein folding energy landscape
Abstract
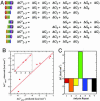
Energy landscapes have been used to conceptually describe and model protein folding but have been difficult to measure experimentally, in large part because of the myriad of partly folded protein conformations that cannot be isolated and thermodynamically characterized. Here we experimentally determine a detailed energy landscape for protein folding. We generated a series of overlapping constructs containing subsets of the seven ankyrin repeats of the Drosophila Notch receptor, a protein domain whose linear arrangement of modular structural units can be fragmented without disrupting structure. To a good approximation, stabilities of each construct can be described as a sum of energy terms associated with each repeat. The magnitude of each energy term indicates that each repeat is intrinsically unstable but is strongly stabilized by interactions with its nearest neighbors. These linear energy terms define an equilibrium free energy landscape, which shows an early free energy barrier and suggests preferred low-energy routes for folding.
Figures

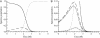
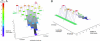
References
-
- Matthews, C. R. (1993) Annu. Rev. Biochem. 62, 653–683. - PubMed
-
- Socci, N. D., Onuchic, J. N. & Wolynes, P. G. (1998) Proteins 32, 136–158. - PubMed
-
- Onuchic, J. N., Nymeyer, H., Garcia, A. E., Chahine, J. & Socci, N. D. (2000) Adv. Protein. Chem. 53, 87–152. - PubMed
-
- Veitshans, T., Klimov, D. & Thirumalai, D. (1997) Folding Des. 2, 1–22. - PubMed
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

