Transmission cycles, host range, evolution and emergence of arboviral disease
- PMID: 15378043
- PMCID: PMC7097645
- DOI: 10.1038/nrmicro1006
Transmission cycles, host range, evolution and emergence of arboviral disease
Abstract
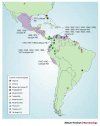
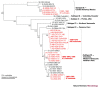
Many pandemics have been attributed to the ability of some RNA viruses to change their host range to include humans. Here, we review the mechanisms of disease emergence that are related to the host-range specificity of selected mosquito-borne alphaviruses and flaviviruses. We discuss viruses of medical importance, including Venezuelan equine and Japanese encephalitis viruses, dengue viruses and West Nile viruses.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

