Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein
- PMID: 15379550
- PMCID: PMC3670950
- DOI: 10.1021/bi0494166
Fetal Alz-50 clone 1 interacts with the human orthologue of the Kelch-like Ech-associated protein
Abstract
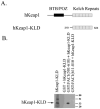
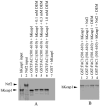
The fetal Alz-50 reactive clone 1 (FAC1) protein exhibits altered expression and subcellular localization during neuronal development and neurodegenerative diseases such as Alzheimer's disease. Using the yeast two-hybrid screen, the human orthologue of Keap1 (hKeap1) was identified as a FAC1 interacting protein. Keap1 is an important regulator of the oxidative stress response pathway through its interaction with the Nrf family of transcription factors. An interaction between full-length FAC1 and hKeap1 proteins has been demonstrated, and the FAC1 binding domain of hKeap1 has been identified as the Kelch repeats. In addition, FAC1 colocalizes with endogenous Keap1 within the cytoplasm of PT67 cells. Exogenously introduced eGFP:hKeap1 fusion protein redistributed FAC1 to colocalize with eGFP:hKeap1 in perinuclear, spherical structures. The interaction between FAC1 and hKeap1 is reduced by competition with the Nrf2 protein. However, competition by Nrf2 for hKeap1 is reduced by diethylmaleate (DEM), a known disrupter of the Nrf2:Keap1 interaction. DEM does not affect the ability of FAC1 to bind hKeap1 in our assay. These results suggest that hKeap1 regulates FAC1 in addition to its known role in control of Nrf2. Furthermore, the observed competition between FAC1 and Nrf2 for binding hKeap1 indicates that the interplay between these three proteins has important implications for neuronal response to oxidative stress.
Figures




References
-
- Jordan-Sciutto KL, Dragich JM, Bowser R. DNA binding activity of the fetal Alz-50 clone 1 (FAC1) protein is enhanced by phosphorylation. Biochem Biophys Res Commun. 1999;260:785–789. - PubMed
-
- Styren SD, Bowser R, Dekosky ST. Expression of fetal ALZ-50 reactive clone 1 (FAC1) in dentate gyrus following entorhinal cortex lesion. J Comp Neurol. 1997;386:555–561. - PubMed
-
- Mu X, Springer JE, Bowser R. FAC1 expression and localization in motor neurons of developing, adult, and amyotrophic lateral sclerosis spinal cord. Exp Neurol. 1997;146:17–24. - PubMed
-
- Schoonover S, Davies P, Bowser R. Immunolocalization and redistribution of the FAC1 protein in Alzheimer's disease. J Neuropathol Exp Neurol. 1996;55:444–455. - PubMed
-
- Bowser R, Giambrone A, Davies P. FAC1, a novel gene identified with the monoclonal antibody Alz50, is develop-mentally regulated in human brain. Dev Neurosci. 1995;17:20–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

